Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 14 July 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools, including a number of preprints on generative AI.
SpheroScan: A User-Friendly Deep Learning Tool for Spheroid Image Analysis
Akshay Akshay, Mitali Katoch, Masoud Abedi, Mustafa Besic, Navid Shekarchizadeh, Fiona C. Burkhard, Alex Bigger-Allen, Rosalyn M. Adam, Katia Monastyrskaya, Ali Hashemi Gheinani
SynBot: An open-source image analysis software for automated quantification of synapses
Justin T. Savage, Juan Ramirez, W. Christopher Risher, Dolores Irala, Cagla Eroglu
Independent regulation of Z-lines and M-lines during sarcomere assembly in cardiac myocytes revealed by the automatic image analysis software sarcApp
Abigail C. Neininger-Castro, James B. Hayes Jr., Zachary C. Sanchez, Nilay Taneja, Aidan M. Fenix, Satish Moparthi, Stéphane Vassilopoulos, Dylan T. Burnette
Integrating Image and Molecular Profiles for Spatial Transcriptomics Analysis
Xi Jiang, Shidan Wang, Lei Guo, Zhuoyu Wen, Liwei Jia, Lin Xu, Guanghua Xiao, Qiwei Li
RNA-to-image multi-cancer synthesis using cascaded diffusion models
Francisco Carrillo-Perez, Marija Pizurica, Yuanning Zheng, Tarak Nath Nandi, Ravi Madduri, Jeanne Shen, Olivier Gevaert
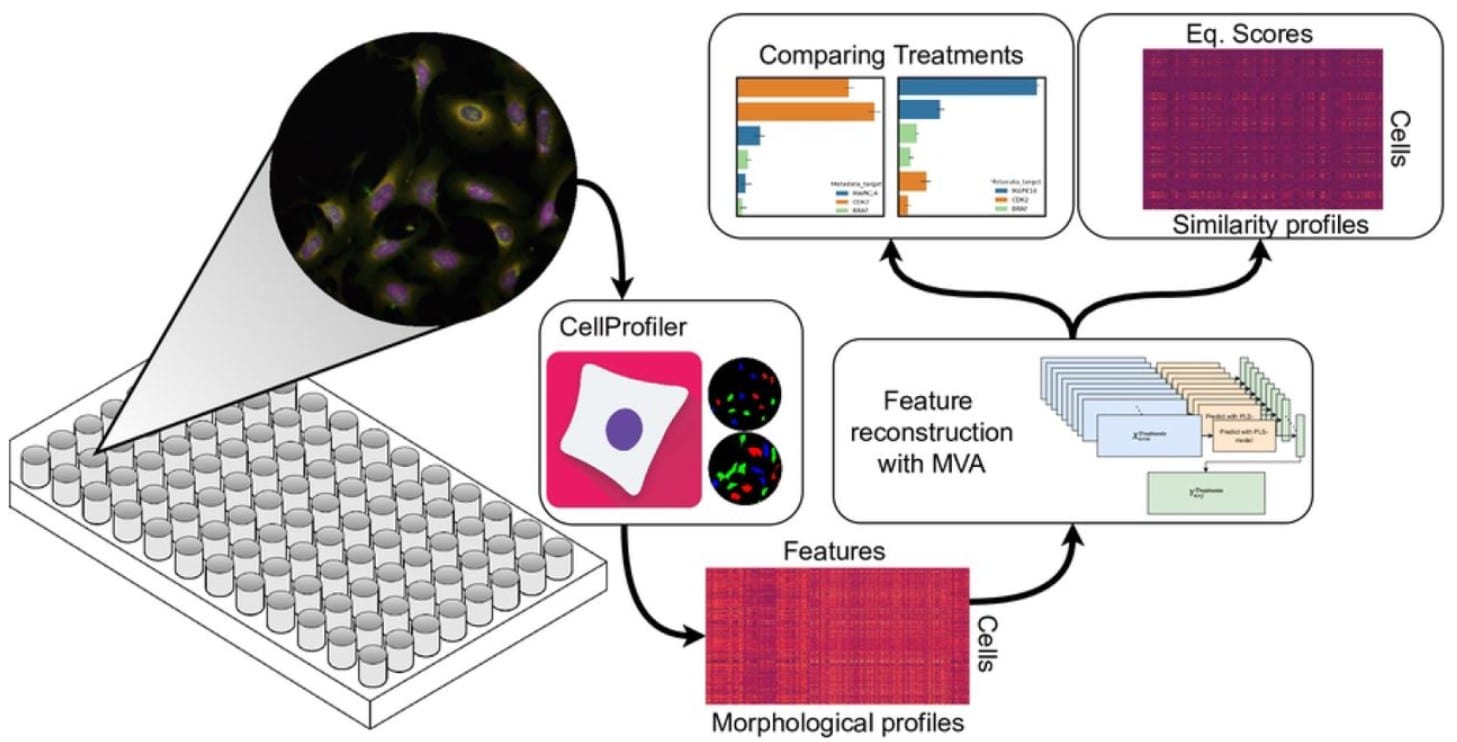
A scalable, data analytics workflow for image-based morphological profiles
Edvin Forsgren, Olivier Cloarec, Pär Jonsson, Johan Trygg
CellStitch: 3D Cellular Anisotropic Image Segmentation via Optimal Transport
Yining Liu, Yinuo Jin, Elham Azizi, Andrew J. Blumberg
An error correction strategy for image reconstruction by DNA sequencing microscopy
Alexander M Kloosterman, Igor Baars, Bjorn Hogberg
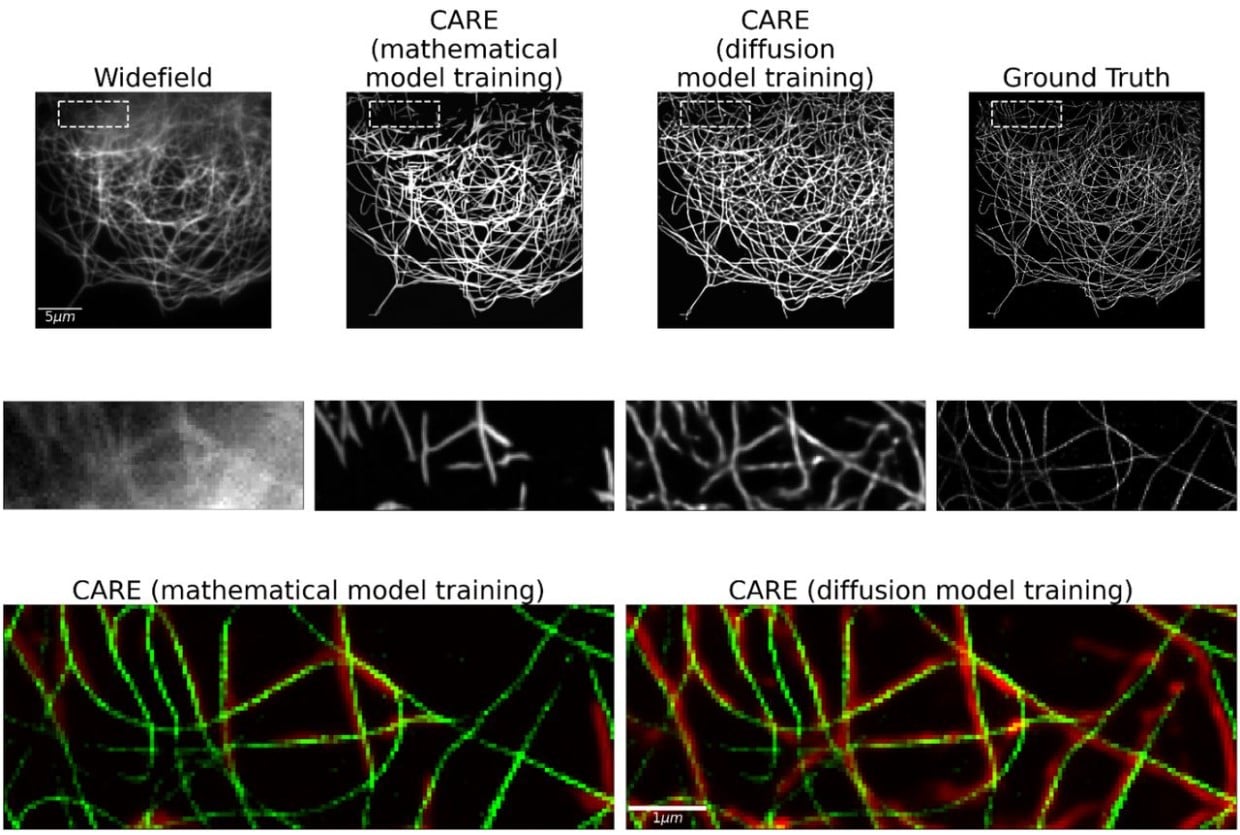
This microtubule does not exist: Super-resolution microscopy image generation by a diffusion model
Alon Saguy, Tav Nahimov, Maia Lehrman, Onit Alalouf, Yoav Shechtman
Generation of Super-resolution Images from Barcode-based Spatial Transcriptomics Using Deep Image Prior
Jeongbin Park, Seungho Cook, Dongjoo Lee, Jinyeong Choi, Seongjin Yoo, Hyung-Jun Im, Daeseung Lee, Hongyoon Choi
Tutorial: guidelines for manual cell type annotation of single-cell multi-omics datasets using interactive software
Yang Joon Kim, Alexander Tarashansky, Karen Liang, Meghan Urisko, Leah Dorman, Michael Borja, Norma Neff, Angela Oliveira Pisco, Alejandro Granados
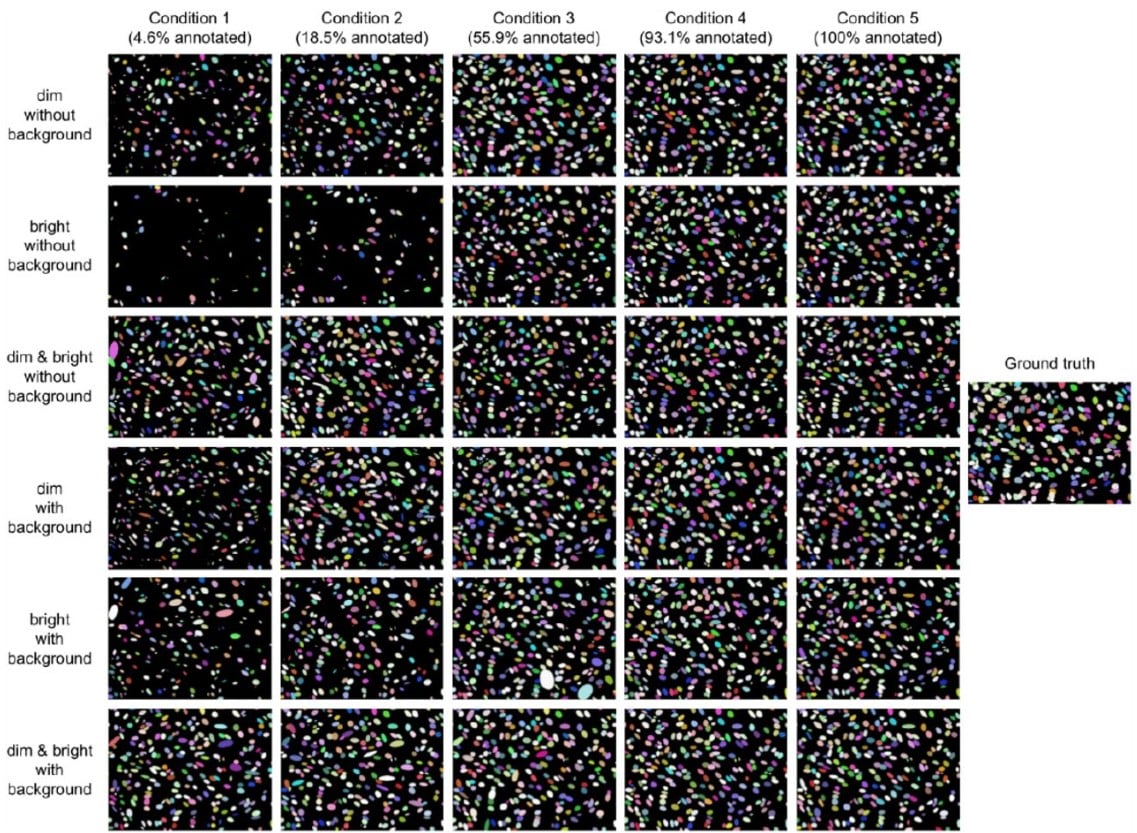
Training deep learning models for cell image segmentation with sparse annotations
Ko Sugawara
Extended correlation functions for spatial analysis of multiplex imaging data
Joshua A. Bull, Eoghan J. Mulholland, Simon J. Leedham, Helen M. Byrne
Tools for Quantitative Analysis of Calcium Signaling Data Using Jupyter-Lab Notebooks
John Rugis, James Chaffer, James Sneyd, David Yule
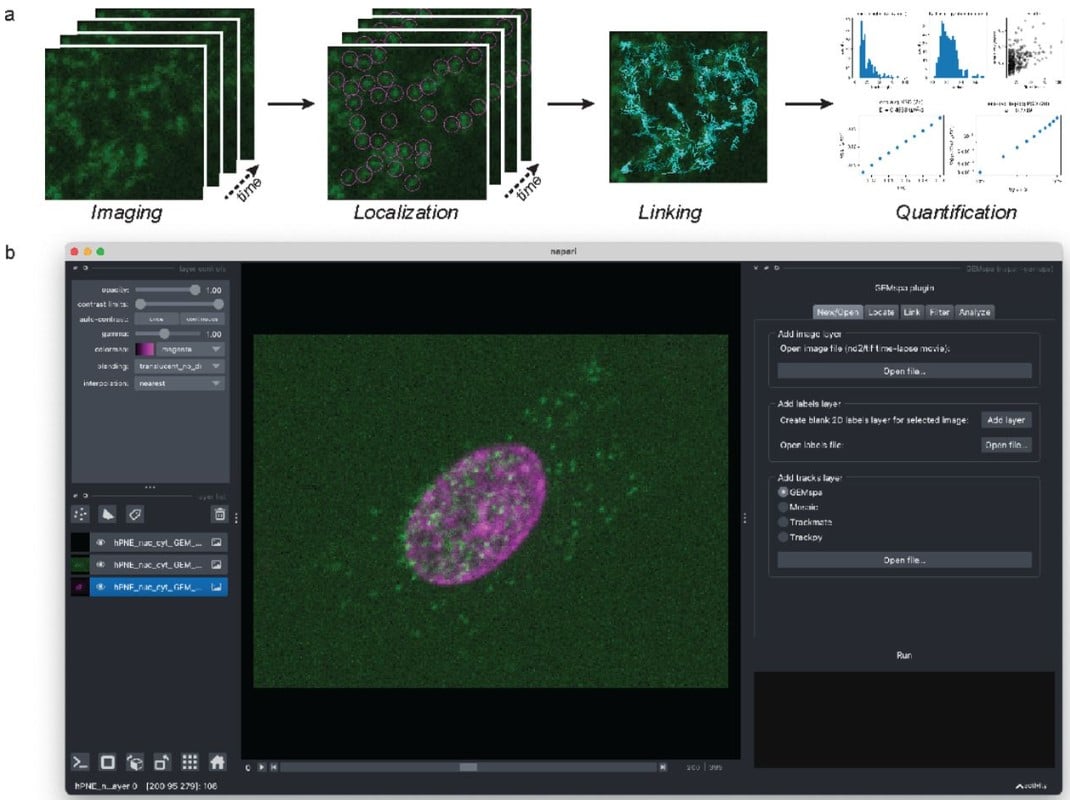
GEMspa: a Napari plugin for analysis of single particle tracking data
Sarah Keegan, David Fenyö, Liam J. Holt
Bridging Imaging Users to Imaging Analysis – A community survey
Suganya Sivagurunathan, Stefania Marcotti, Carl J Nelson, Martin L Jones, David J Barry, Thomas J A Slater, Kevin W Eliceiri, Beth A Cimini
JDLL: A library to run Deep Learning models on Java bioimage informatics platforms
Carlos Garcia Lopez de Haro, Stephane Dallongeville, Thomas Musset, Estibaliz Gomez de Mariscal, Daniel Sage, Wei Ouyang, Arrate Munoz-Barrutia, Jean-Yves Tinevez, Jean-Christophe Olivo-Marin
ModularImageAnalysis (MIA): Assembly of modularised image and object analysis workflows in ImageJ
Stephen J. Cross, Jordan D. J. R. Fisher, Mark A. Jepson
Virtual tissue microstructure reconstruction by deep learning and fluorescence microscopy
Nicolás Bettancourt, Cristian Pérez, Valeria Candia, Pamela Guevara, Yannis Kalaidzidis, Marino Zerial, Fabián Segovia-Miranda, Hernán Morales-Navarrete
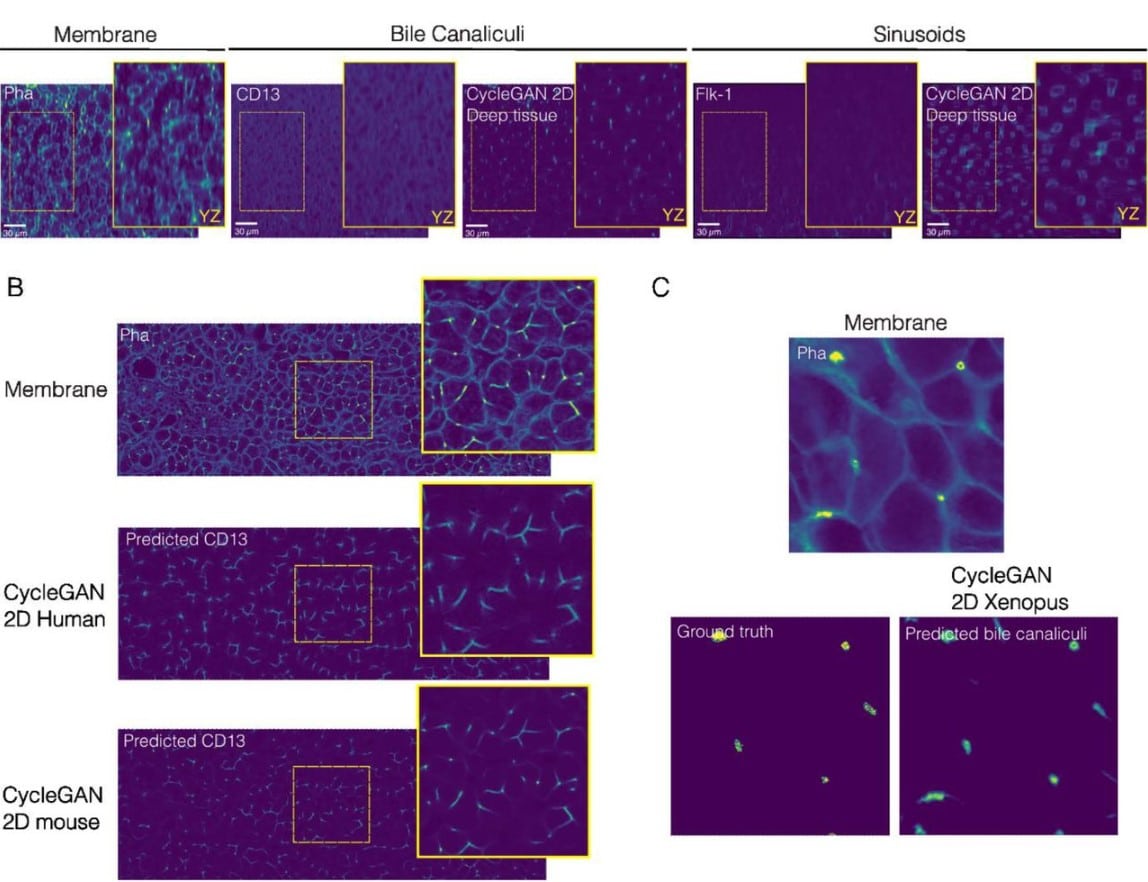
Figure extracted from Bettancourt, et al.
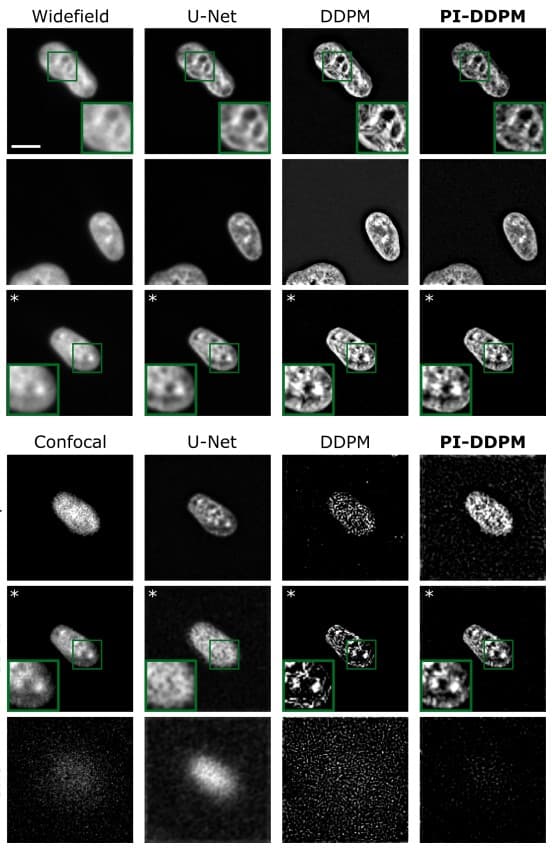
Microscopy image reconstruction with physics-informed denoising diffusion probabilistic model
Rui Li, Gabriel della Maggiora, Vardan Andriasyan, Anthony Petkidis, Artsemi Yushkevich, Mikhail Kudryashev, Artur Yakimovich


 (No Ratings Yet)
(No Ratings Yet)