Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 7 March 2025
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
Parameter Efficient Fine-Tuning of Segment Anything Model
Carolin Teuber, Anwai Archit, Constantin Pape
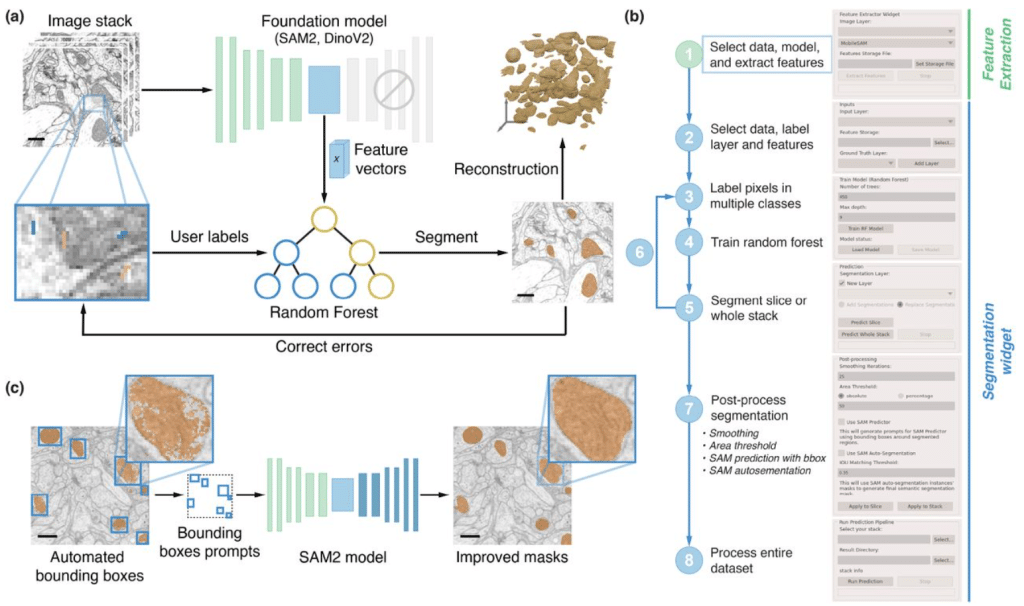
FeatureForest: the power of foundation models, the usability of random forests
Mehdi Seifi, Damian Dalle Nogare, Juan Battagliotti, Vera Galinova, Ananya Kedige Rao, AI4Life Horizon Europe Programme Consortium, Johan Decelle, Florian Jug, Joran Deschamps
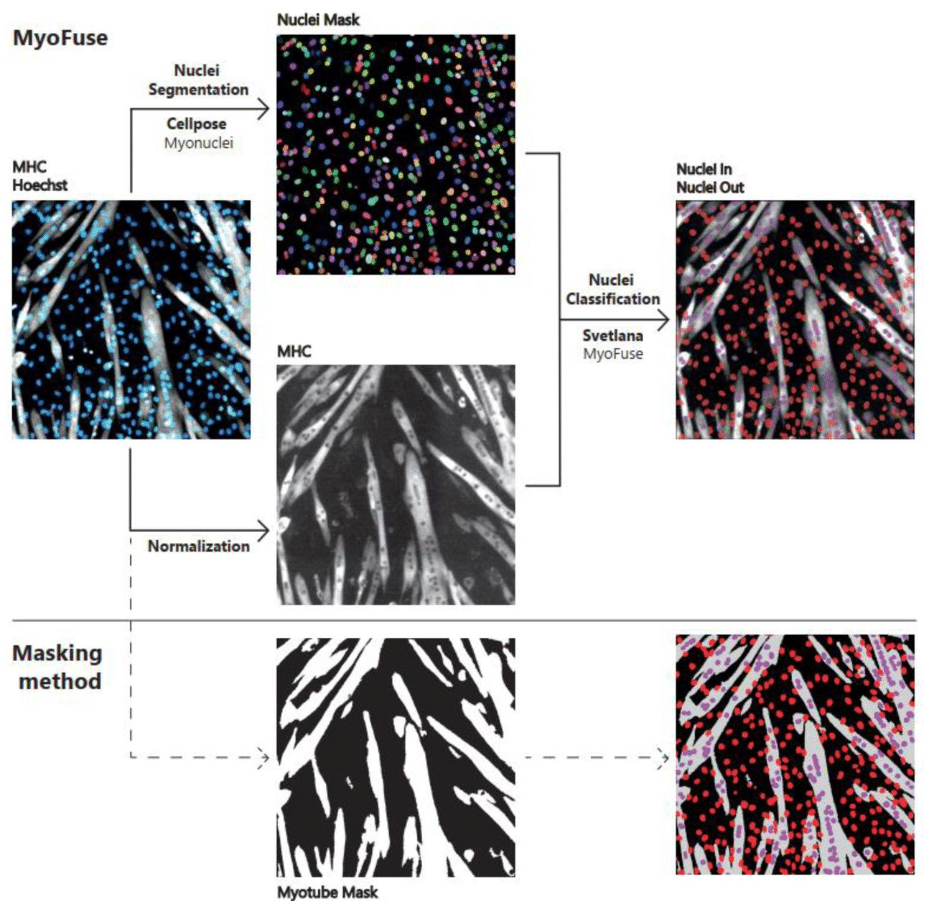
MyoFuse: A fully AI-based workflow for automated quantification of skeletal muscle cell fusion in vitro
Benjamin Lair, Clément Cazorla, Alicia Lobeto, Axel Labour, Claire Laurens, Arild C. Rustan, Pierre Weiss, Remy Flores-Flores, Cedric Moro
A deep learning-based noise correction method for light-field fluorescence microscopy
Bohan Qu, Zhouyu Jin, You Zhou, Bo Xiong, Xun Cao
ProPicker: Promptable Segmentation for Particle Picking in Cryogenic Electron Tomography
Simon Wiedemann, Zalan Fabian, Mahdi Soltanolkotabi, Reinhard Heckel
An Open-Source Image Analysis Method for Quantifying Reporter Fluorescence
Kathryn F. Ball, Jacob A. Perez, Allen M. Cooper, Charmaine U. Pira, Kerby C. Oberg, Christopher G. Wilson

MassVision: An open-source end-to-end platform for AI-driven mass spectrometry image analysis
Amoon Jamzad, Ayesha Syeda, Jade Warren, Martin Kaufmann, Natasha Iaboni, Christopher J. B. Nicol, John Rudan, Kevin Y. M. Ren, David Hurlbut, Sonal Varma, Gabor Fichtinger, Parvin Mousavi
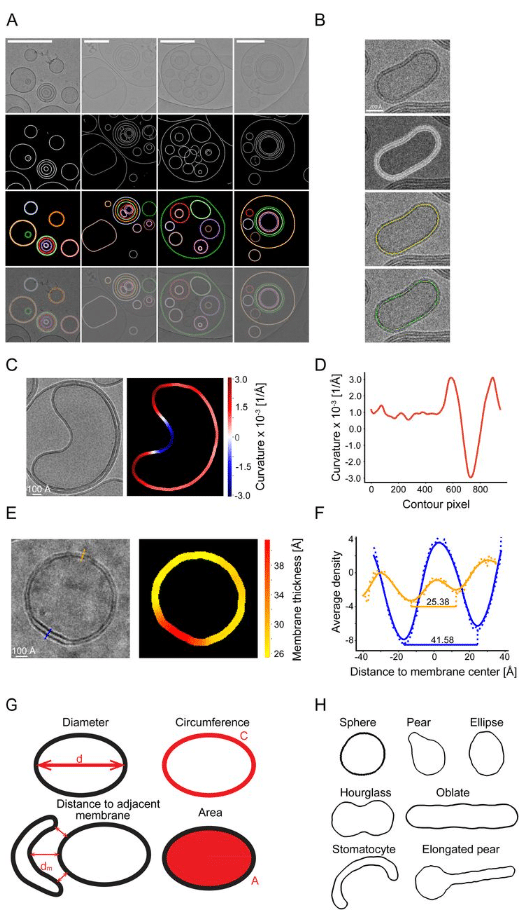
CryoVIA – An image analysis toolkit for the quantification of membrane structures from cryo-EM micrographs
Philipp Schönnenbeck, Benedikt Junglas, Carsten Sachse
LEVERAGING TRANSFER LEARNING FOR HIGH-ACCURACY PHENOTYPIC SCREENING IN ZEBRAFISH IMAGE ANALYSIS
Varshini Uma Jayaraman, Raghavender Medishetti, Shreetama Ghosh, Kiranam Chatti, Mahesh Kumar Uppada, Srinivas Oruganti
stDyer-image improves clustering analysis of spatially resolved transcriptomics and proteomics with morphological images
Ke Xu, Xin Maizie Zhou, Lu Zhang
Developing the Computational Image Processing Method for Quantitative Analysis of Nanopore Structure Obtained from HS-AFM (AFMnanoQ)
Thuyen Vinh Tran Nguyen, Ngoc Quoc Ly, Ngan Thi Phuong Le, Hoang Duc Nguyen, Kien Xuan Ngo
Self-supervised image restoration in coherent X-ray neuronal microscopy
Alfred Laugros, Peter Cloetens, Carles Bosch, Richard Schoonhoven, Liam Pavlovic, Aaron T. Kuan, Jayde Livingstone, Yuxin Zhang, Minsu Kim, Allard Hendriksen, Mirko Holler, Adrian A. Wanner, Anthony Azevedo, K. Joost Batenburg, John C. Tuthill, Wei-Chung Allen Lee, Andreas T. Schaefer, Nicola Vigano, Alexandra Pacureanu
ViFIT-assisted Histopathology: From H&E Style Standardization to Virtual Fiber Image Transformation
Shu Wang, Xiao Zhang, Xingfu Wang, Chenyong Lv, Xiahui Han, Xiong Lin, Deyong Kang, Ruolan Lin, Liwen Hu, Feng Huang, Wenxi Liu, Jianxin Chen
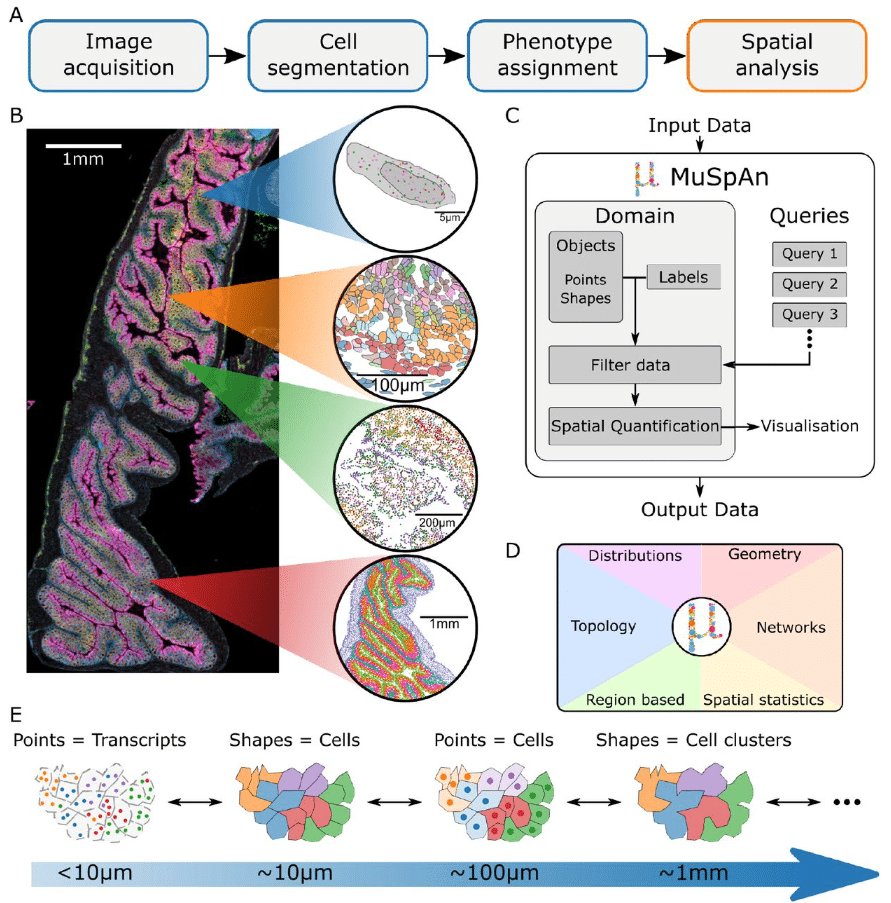
MuSpAn: A Toolbox for Multiscale Spatial Analysis
Joshua A. Bull, Joshua W. Moore, Eoghan J. Mulholland, Simon J. Leedham, Helen M. Byrne
Enhanced fluorescence lifetime imaging microscopy denoising via principal component analysis
Soheil Soltani, Jack G. Paulson, Emma J. Fong, Shannon M. Mumenthaler, Andrea M. Armani
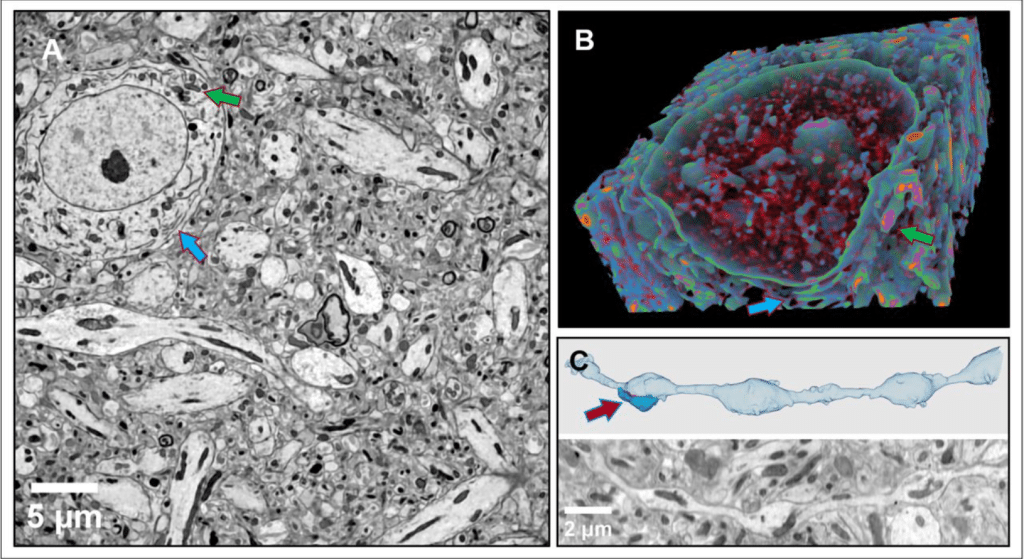
Semi-automated analysis of beading in degenerating axons
V C Pretheesh Kumar, Pramod Pullarkat
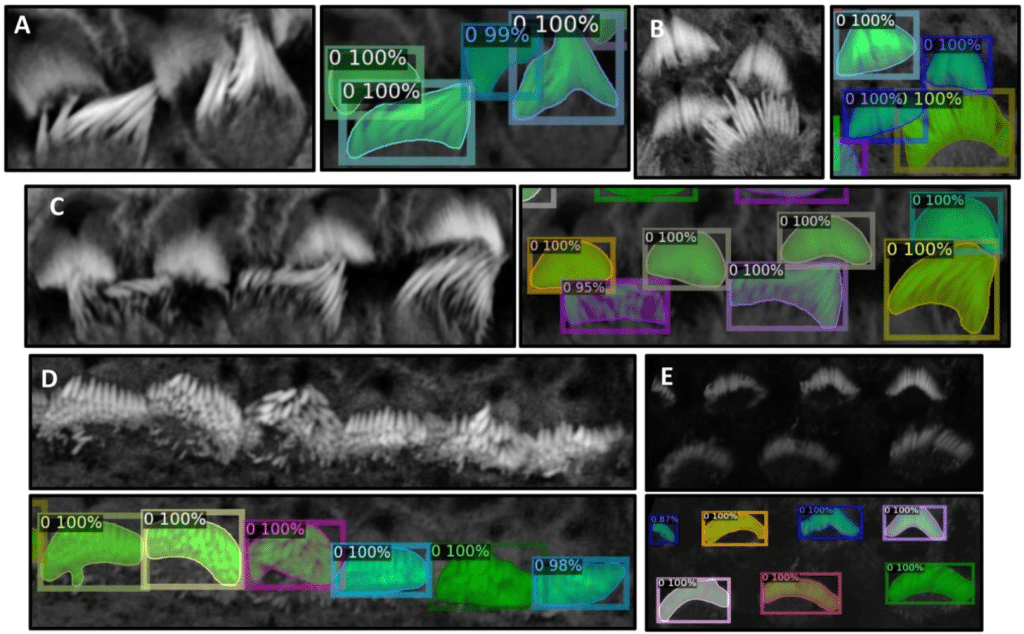
VASCilia (Vision Analysis StereoCilia): A Napari Plugin for Deep Learning-Based 3D Analysis of Cochlear Hair Cell Stereocilia Bundles
Yasmin M. Kassim, David B. Rosenberg, Samprita Das, Zhuoling Huang, Samia Rahman, Ibraheem Al Shammaa, Samer Salim, Kevin Huang, Alma Renero, Cayla Miller, Yuzuru Ninoyu, Rick A. Friedman, Artur Indzhykulian, Uri Manor
GSLab: Open-Source Platform for Advanced Phasor Analysis in Fluorescence Microscopy
Alexander Vallmitjana, Belén Torrado, Amanda F. Durkin, Alexander Dvornikov, Navid Rajil, Suman Ranjit, Mihaela Balu
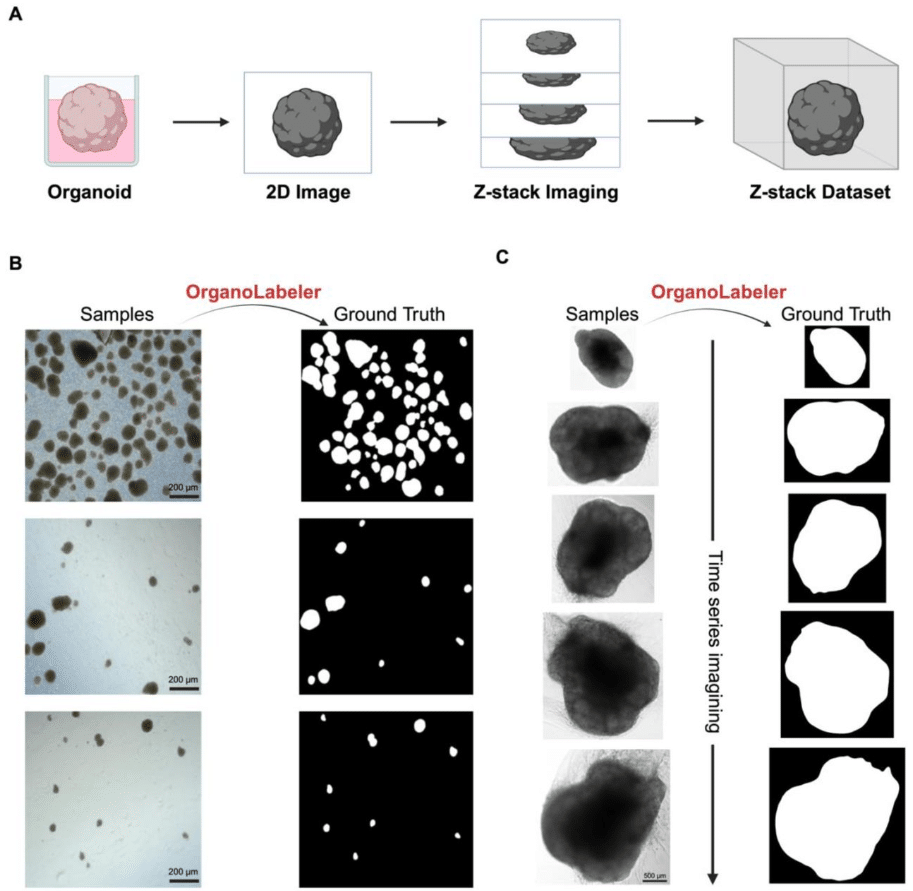
BrAIn: A comprehensive artificial intelligence-based morphology analysis system for brain organoids and neuroscience
Burak Kahveci, Elifsu Polatli, Ali Eren Evranos, Hüseyin Güner, Yalin Bastanlar, Gökhan Karakülah, Sinan Güven
DeepSpaceDB: a spatial transcriptomics atlas for interactive in-depth analysis of tissues and tissue microenvironments
Vladyslav Honcharuk, Afeefa Zainab, Yoshiya Horimoto, Keiko Takemoto, Diego Diez, Shinpei Kawaoka, Alexis Vandenbon
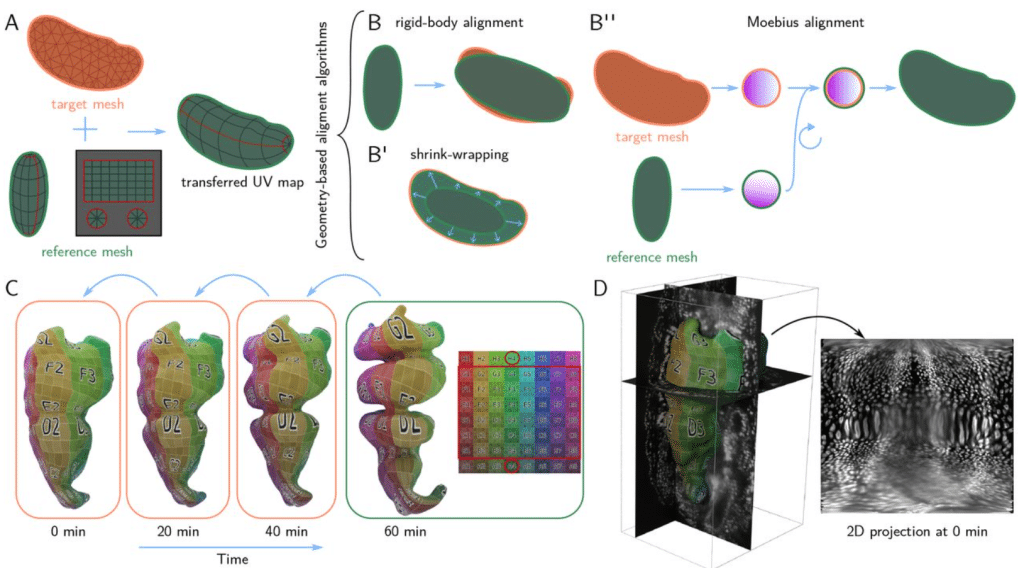
Blender tissue cartography: an intuitive tool for the analysis of dynamic 3D microscopy data
Nikolas Claussen, Cécile Regis, Susan Wopat, Sebastian Streichan
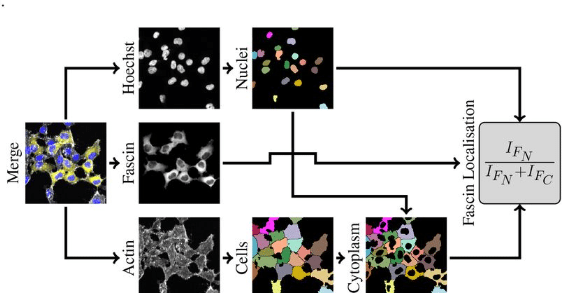
Enhancing Reproducibility Through Bioimage Analysis: The Significance of Effect Sizes and Controls
David J. Barry, Stefania Marcotti, Lina Gerontogianni, Gavin Kelly
SPTnet: a deep learning framework for end-to-end single-particle tracking and motion dynamics analysis
Cheng Bi, Kevin L. Scrudders, Yue Zheng, Maryam Mahmoodi, Shalini T. Low-Nam, Fang Huang
LivecellX: A Deep-learning-based, Single-Cell Object-Oriented Framework for Quantitative Analysis in Live-Cell Imaging
Ke Ni, Gaohan Yu, Zhiqian Zheng, Yong Lu, Dante Poe, Shiman Zhou, Weikang Wang, Jianhua Xing
CalciumNetExploreR: An R Package for Network Analysis of Calcium Imaging Data
Simone Lenci, Dirk Sieger


 (No Ratings Yet)
(No Ratings Yet)