Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 22 August 2025
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
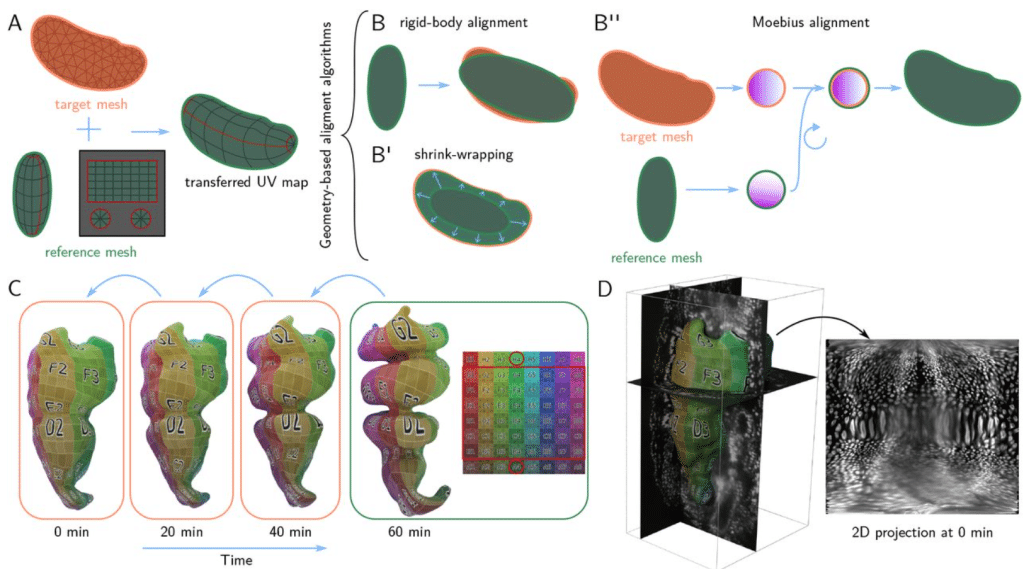
Blender tissue cartography: an intuitive tool for the analysis of dynamic 3D microscopy data
Nikolas Claussen, Cécile Regis, Susan Wopat, Sebastian Streichan
SEAL: Spatially-resolved Embedding Analysis with Linked Imaging Data
Simon Warchol, Grace Guo, Johannes Knittel, Dan Freeman, Usha Bhalla, Jeremy L Muhlich, Peter K. Sorger, Hanspeter Pfister
High-content high-resolution microscopy and deep learning assisted analysis reveals host and bacterial heterogeneity during Shigella infection
Ana T. López-Jiménez, Dominik Brokatzky, Kamla Pillay, Tyrese Williams, Gizem Özbaykal Güler, Serge Mostowy
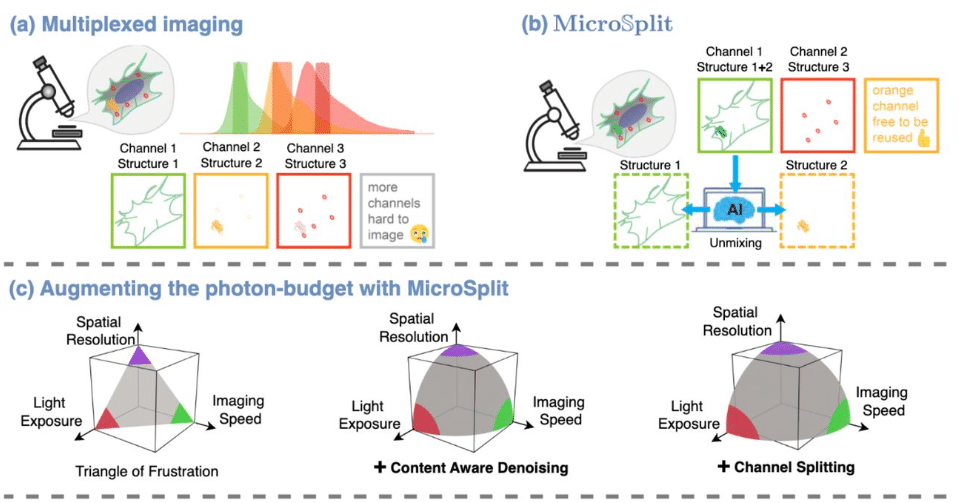
Micro𝕊plit: Semantic Unmixing of Fluorescent Microscopy Data
Ashesh Ashesh, Federico Carrara, Igor Zubarev, Vera Galinova, Melisande Croft, Melissa Pezzotti, Daozheng Gong, Francesca Casagrande, Elisa Colombo, Stefania Giussani, Elena Restelli, Eugenia Cammarota, Juan Manuel Battagliotti, Nikolai Klena, Moises Di Sante, Raghabendra Adhikari, Daniel Feliciano, Gaia Pigino, Elena Taverna, Oliver Harschnitz, Nicola Maghelli, Norbert Scherer, Damian Edward Dalle Nogare, Joran Deschamps, Francesco Pasqualini, Florian Jug
Deep learning-based image quantification of epithelial cell shapes and its application to polycystic kidney disease
Johannes Jahn, Alexis Hofherr, Clara Consoli, Berenike Fajen, Rebekka Goll, Greta Theresa Liedtke, Adrian Boehm, Friederike Selbach, Paul Christoph Zeisler, Vanessa Weichselberger, Anne-Kathrin Classen, Lukas Westermann, Tilman Busch, Michael Köttgen
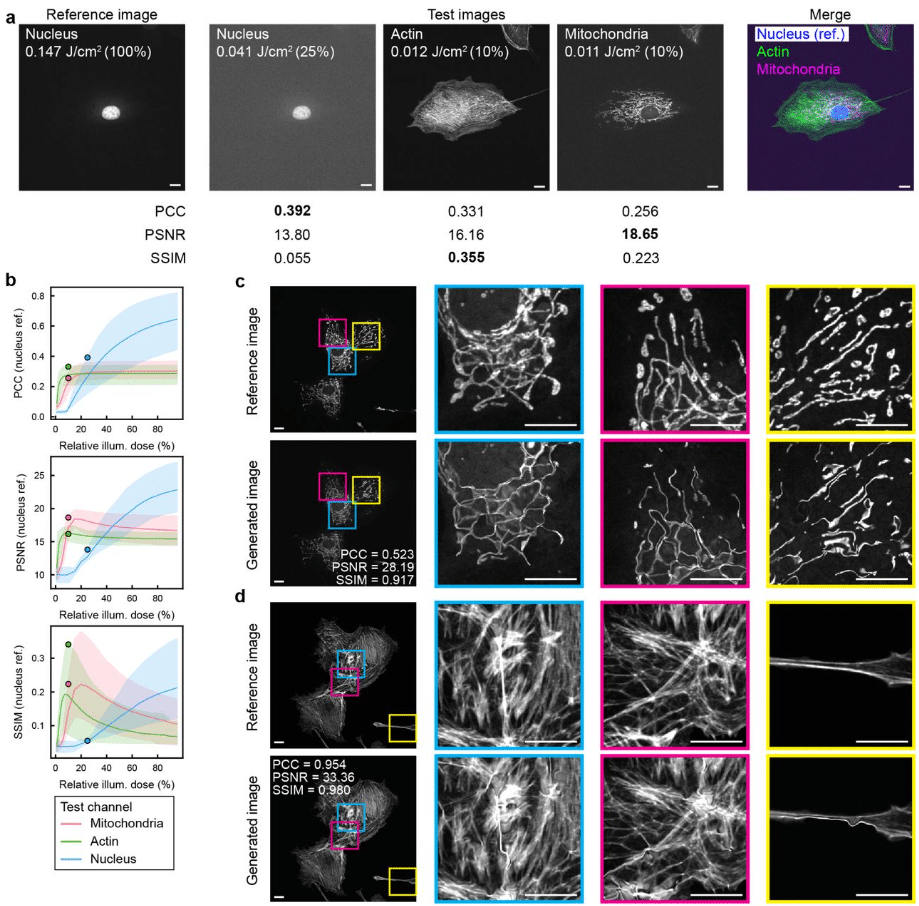
Image quality metrics fail to accurately represent biological information in fluorescence microscopy
Ihuan Gunawan, Richard J Marsh, Nandini Aggarwal, Erik Meijering, Susan Cox, John G Lock, Siân Culley
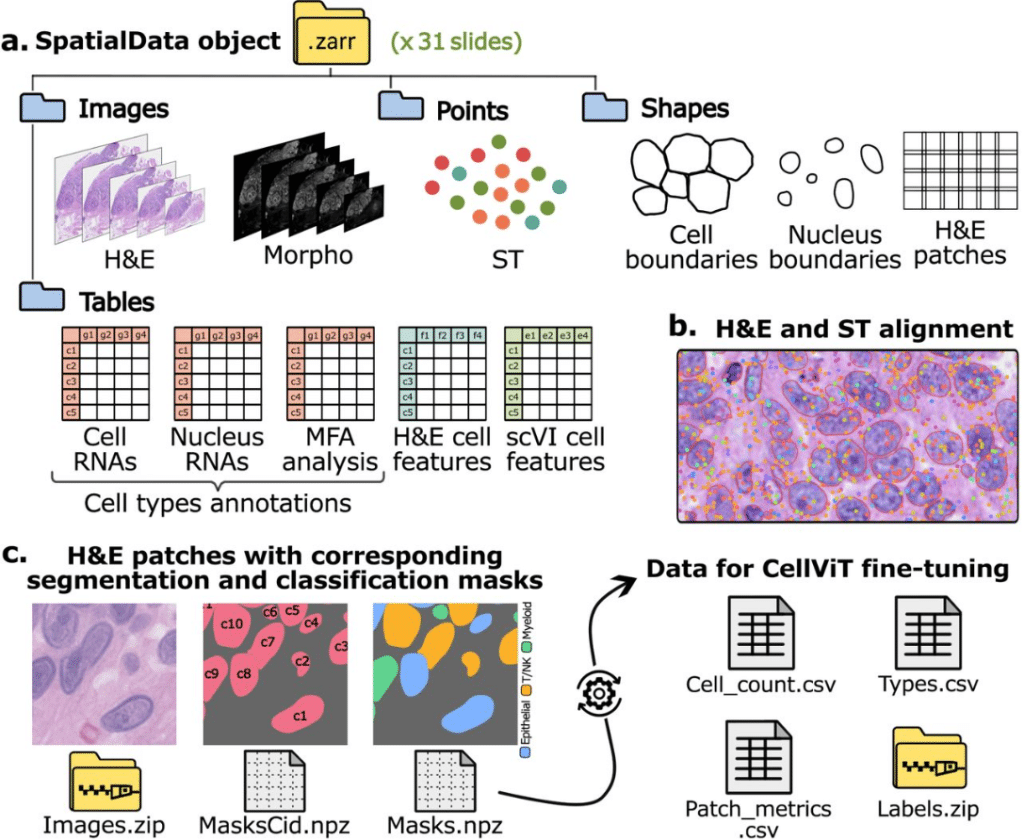
STHELAR, a multi-tissue dataset linking spatial transcriptomics and histology for cell type annotation
Félicie Giraud-Sauveur, Quentin Blampey, Hakim Benkirane, Arianna Marinello, Paul-Henry Cournède, Stergios Christodoulidis
SPROUT: A User-friendly, Scalable Toolkit for Multi-class Segmentation of Volumetric Images
Yichen He, Marco Camaiti, Lucy E. Roberts, James M. Mulqueeney, Eleftherios Ioannou, Marius Didziokas, Anjali Goswami
MiroSCOPE: An AI-driven digital pathology platform for annotating functional tissue units
Madeleine R. Fenner, Selim Sevim, Guanming Wu, Deidre Beavers, Pengfei Guo, Yucheng Tang, Christopher Z. Eddy, Kaoutar Ait-Ahmad, Travis Rice-Stitt, George Thomas, M.J. Kuykendall, Vasilis Stavrinides, Mark Emberton, Daguang Xu, Xubo Song, S. Ece Eksi, Emek Demir
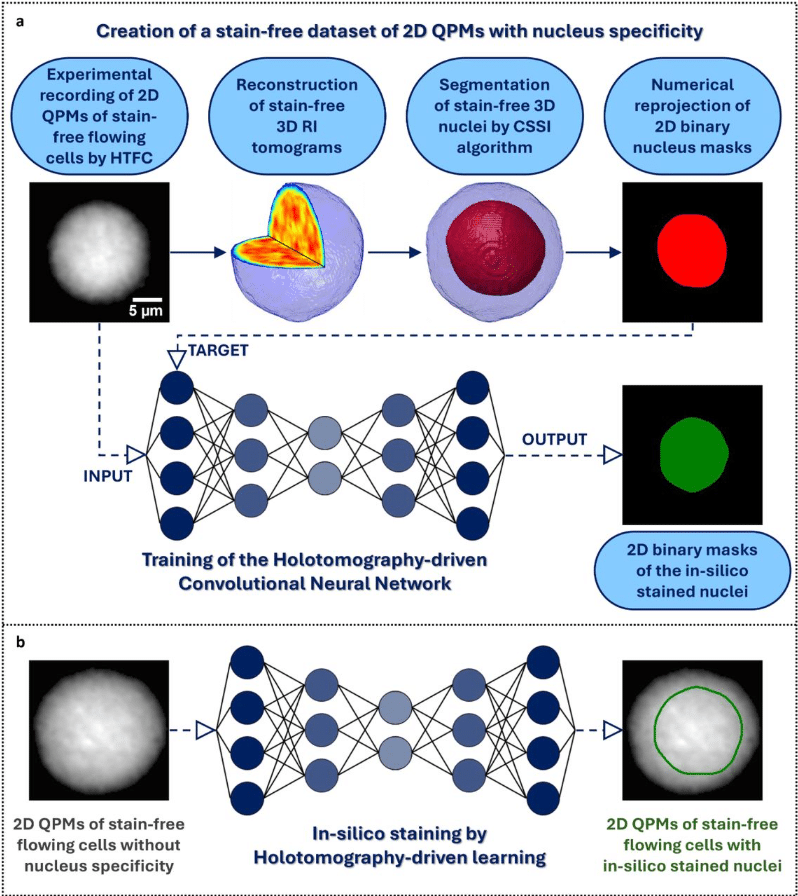
Holotomography-driven learning for in-silico staining of single cells in flow cytometry avoiding co-registration
Daniele Pirone, Giusy Giugliano, Michela Schiavo, Annalaura Montella, Martina Mugnano, Vincenza Cerbone, Maddalena Raia, Giulia Scalia, Ivana Kurelac, Diego Luis Medina, Lisa Miccio, Mario Capasso, Achille Iolascon, Pasquale Memmolo, Pietro Ferraro
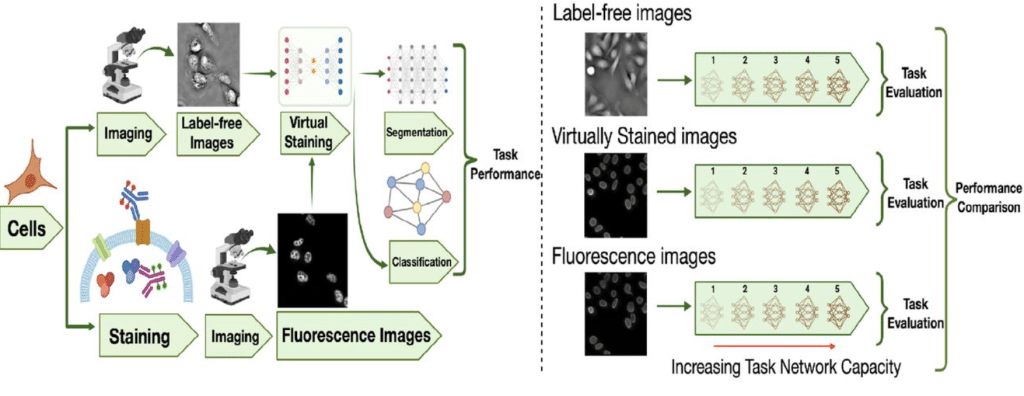
On the Utility of Virtual Staining for Downstream Applications as it relates to Task Network Capacity
Sourya Sengupta, Jianquan Xu, Phuong Nguyen, Frank J. Brooks, Yang Liu, Mark A. Anastasio
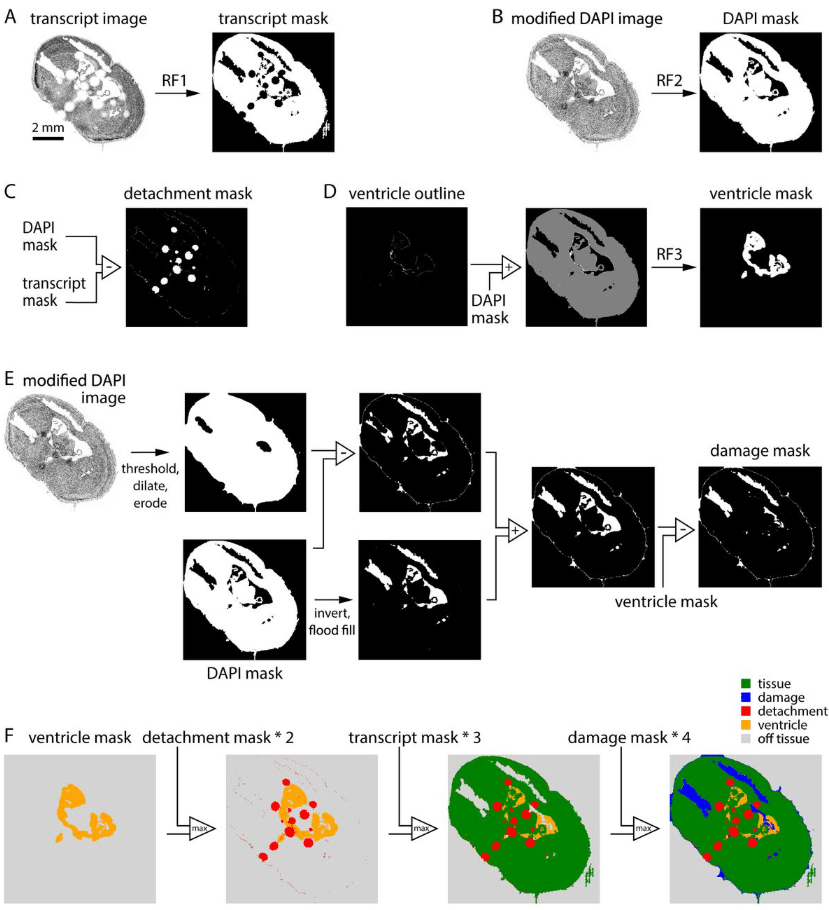
Scalable automated segmentation quantifies mitochondrial proteins and morphology at the nanoscale
Yuan Tian, Alexander Sauer, Mariia Dmitrieva, Hongqing Han, Hannahmariam T. Mekbib, Yujin Bao, Xiaojia Guo, Tian-min Chen, Robert Safirstein, Gary Desir, Jens Rittscher, Joerg Bewersdorf
FlyTomo: A streamlined software for on-the-fly cryo-ET data processing and diagnosis
Zheyuan Zhang, Cheng Peng, Weiping Zhang, Jiaming Liang, Kexin Liu, Yong Chen, Junxia Zhang, Rui Liang, Yutong Song, Sai Li
Anatomy-aware, label-informed approach improves image registration for challenging datasets
Rachel A. Roston, Nicholas J. Tustison, A. Murat Maga
Registration-based 3D Light Sheet Fluorescence Microscopy and 2D histology image fusion tool for pathological specimen
Marcel Brettmacher, Philipp Nolte, Diana Pinkert-Leetsch, Felix Bremmer, Jeannine Missbach-Guentner, Christoph Rußmann
MerQuaCo: a computational tool for quality control in image-based spatial transcriptomics
Naomi Martin, Paul Olsen, Jacob Quon, Jazmin Campos, Nasmil Valera Cuevas, Josh Nagra, Marshall VanNess, Zoe Maltzer, Emily C Gelfand, Alana Oyama, Amanda Gary, Yimin Wang, Angela Alaya, Augustin Ruiz, Cade Reynoldson, Cameron Bielstein, Christina Alice Pom, Cindy Huang, Cliff Slaughterbeck, Elizabeth Liang, Jason Alexander, Jeanelle Ariza, Jocelin Malone, Jose Melchor, Kaity Colbert, Krissy Brouner, Lyudmila Shulga, Melissa Reding, Patrick Latimer, Raymond Sanchez, Stuard Barta, Tom Egdorf, Zachary Madigan, Chelsea M Pagan, Jennie L Close, Brian Long, Michael Kunst, Ed S Lein, Hongkui Zeng, Delissa McMillen, Jack Waters
AI-driven analysis for real-time detection of unstained microscopic cell culture images
Kathrin Hildebrand, Tatiana Mögele, Dennis Raith, Maria Kling, Anna Rubeck, Stefan Schiele, Eelco Meerdink, Avani Sapre, Jonas Bermeitinger, Martin Trepel, Rainer Claus
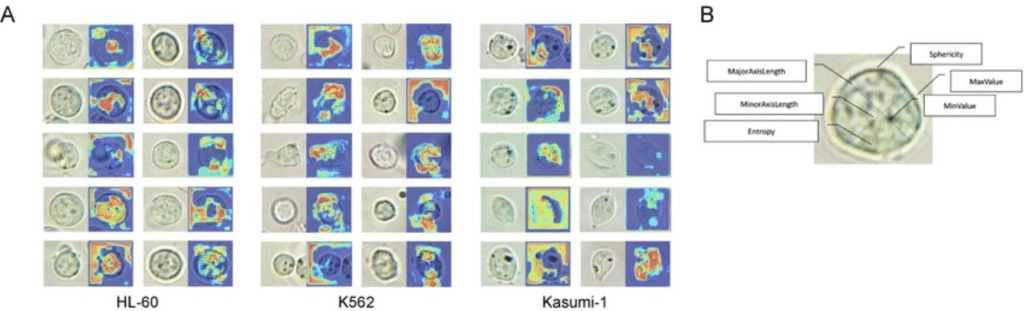
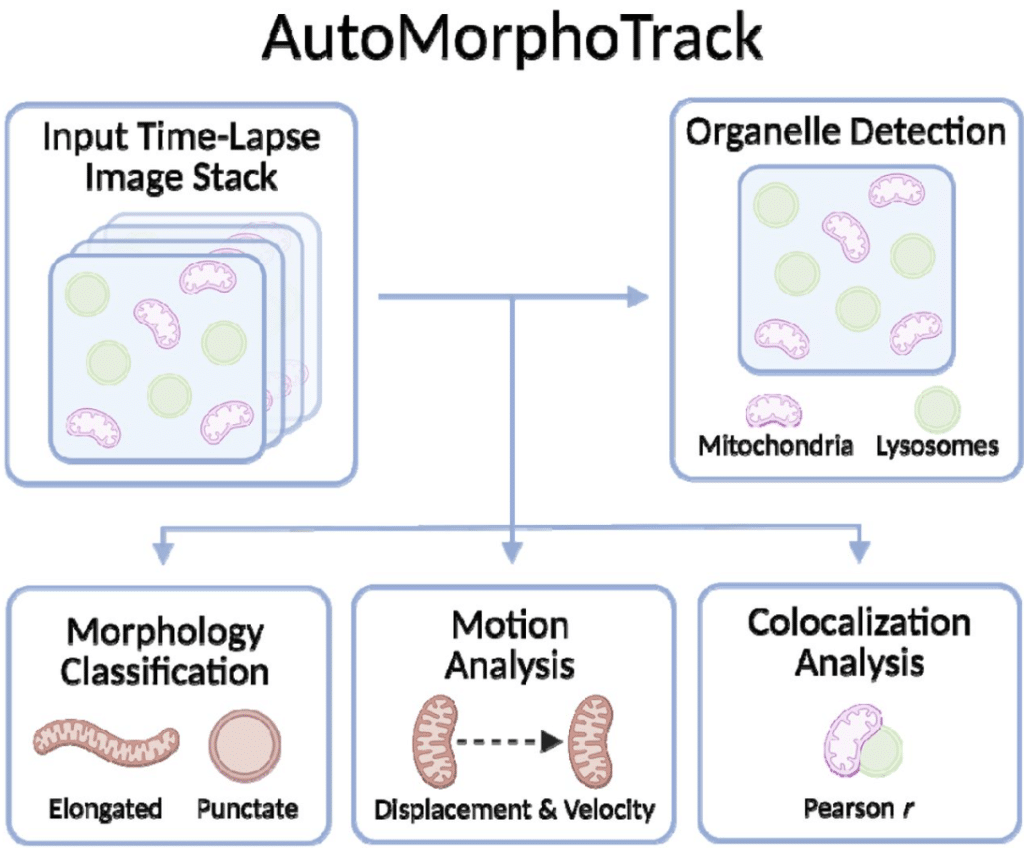
AutoMorphoTrack: An Automated Python Package for Organelle Morphology, Motility, and Colocalization Analysis in Live-Cell Imaging
Armin Bayati, Jackson G. Schumacher, Xiqun Chen
Non-segmented unsupervised learning of multispectral whole slide images for robust analysis of tissue repair and regeneration
Kody Paul Mansfield, Tamara Mestvirishvili, Bibi Sheleeza Subhan, Valeria Mezzano-Robinson, Dianny A Almanzar, Sydney Hanson, Jimin Tan, Cynthia A Loomis, David Fenyo, Aristotelis Tsirigos, Piul S Rabbani
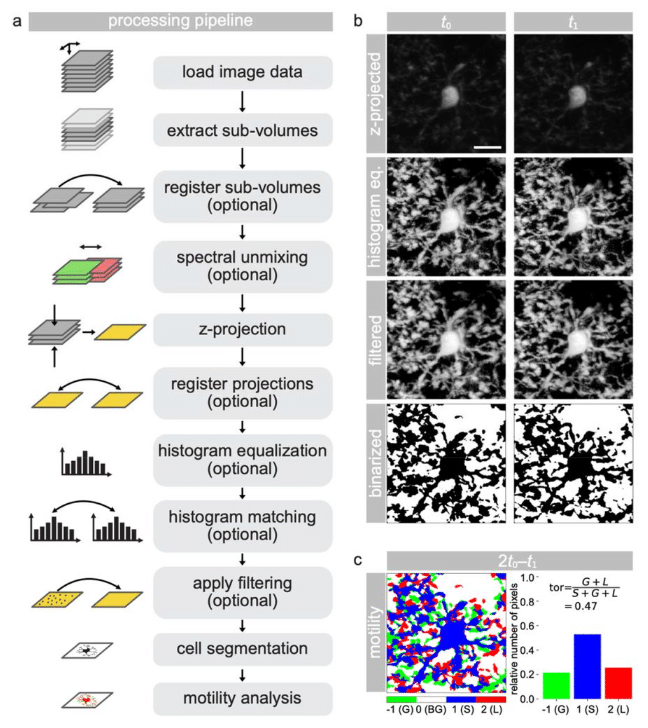
MotilA – A Python pipeline for the analysis of microglial fine process motility in 3D time-lapse multiphoton microscopy data
Fabrizio Musacchio, Sophie Crux, Felix Nebeling, Nala Gockel, Falko Fuhrmann, Martin Fuhrmann
Learned Single-Pixel Fluorescence Microscopy
Serban C. Tudosie, Valerio Gandolfi, Shivaprasad Varakkoth, Andrea Farina, Cosimo D’Andrea, Simon Arridge
Twist and Scout: Analysis and Curation of Particles in Cryo-Electron Tomography Using TANGO
Markus Schreiber, Beata Turoňová
Unsupervised learning of structural variability in cryo-EM data using normal mode analysis of deformable atomic models
Youssef Nashed, Julien Martel, Ariana Peck, Axel Levy, Huanghao Mai, Gordon Wetzstein, Nina Miolane, Daniel Ratner, Frédéric Poitevin
Fast, flexible, learning-free organoid quantification and tracking with OrganoSeg2
Cameron J. Wells, Najwa Labban, Shayna L. Showalter, Róża K. Przanowska, Kevin A. Janes
Quantitative and unbiased lung alveolar septum assessment in a LPS experimental mouse model using 2D-spatial correlation image analysis from hematoxylin and eosin slides
Micaela Lopassio, María José García, Leonel Malacrida
About the alignment and 3D reconstruction of sparse cryo-scanning transmission electron tomography datasets
Sylvain Trépout
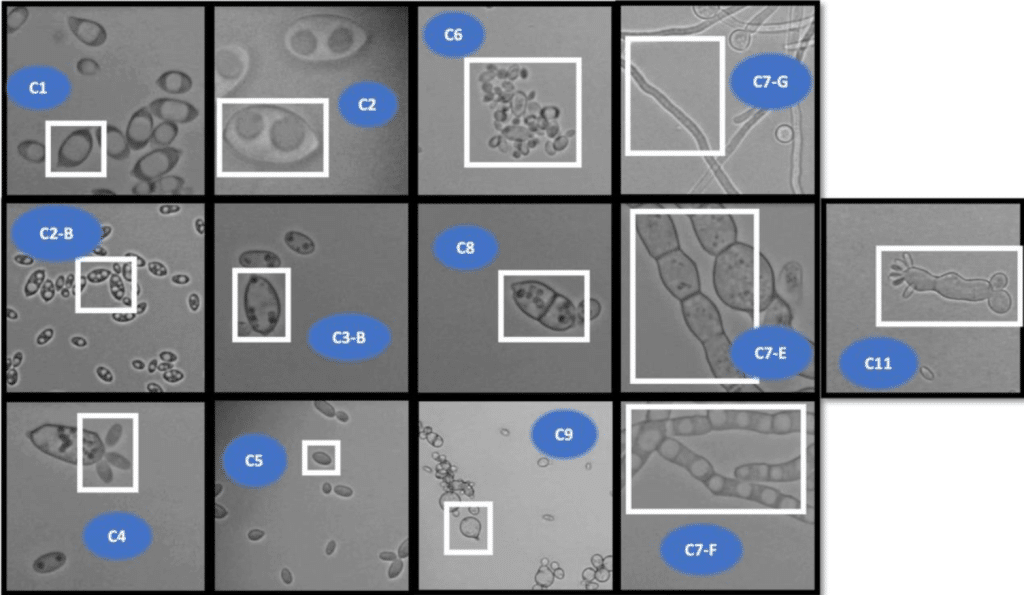
TU_MyCo-Vision: A Deep Learning Tool for Detection of Cell Morphologies in Fungal Microscopic Images
Kartik J. Deopujari, Matthias Schmal, Caroline Danner, Zainab Abdul Qayyum, Jordy T. Zwerus, Julian Kopp, Mihail Besleaga, Roghayeh Shirvani, Astrid R. Mach-Aigner, Robert L. Mach, Christian Zimmermann
DeepPathway: Predicting Pathway Expression from Histopathology Images
Muhammad Ahtazaz Ahsan, Karen Piper Hanley, Martin Fergie, Claire O’leary, Gerben Borst, Federico Roncaroli, Magnus Ratrray, Mudassar Iqbal, Syed Murutza Baker
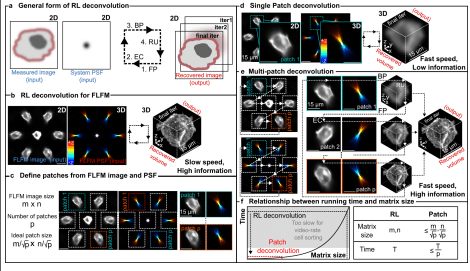
Patch deconvolution for Fourier light-field microscopy
Bin Fu, Caroline L. Jones, Daniel Heraghty, Shengbo Yang, Caitlin O’Brien-Ball, Victoria Junghans, Haowei Yang, Tuomas Knowles, Lucien E. Weiss, Ricardo A. Fernandes, Steven F. Lee
Self-contrastive learning enables interference-resilient and generalizable fluorescence microscopy signal detection without interference modeling
Fengdi Zhang, Ruqi Huang, Meiqian Xin, Haoran Meng, Danheng Gao, Ying Fu, Juntao Gao, Xiangyang Ji
SKOOTS: Skeleton-oriented object segmentation for mitochondria
Christopher J Buswinka, Richard T. Osgood, Hidetomi Nitta, Artur A. Indzhykulian
MitoStructSeg: mitochondrial structural complexity resolution via adaptive learning for cross-sample morphometric profiling
Xinsheng Wang, Xiaohua Wan, Buqing Cai, Zhuo Jia, Yuanbo Chen, Shuai Guo, Zheng Liu, Fuwei Li, Fa Zhang, Bin Hu


 (No Ratings Yet)
(No Ratings Yet)