Microscopy preprints: bioimage analysis
Posted by FocalPlane, on 29 December 2025
Here is a curated selection of preprints published (or updated) recently. In this post we focus specifically on bioimage analysis and data management.
CochleaNet: deep learning-based image analysis for cochlear connectomics and gene therapy
Lennart Roos, Aleyna Miraç Diniz, Elisabeth Koert, Martin Schilling, Mara Uhl, Anupriya Thirumalai, Mostafa Aakhte, Kathrin Kusch, Jan Huisken, Tobias Moser, Constantin Pape
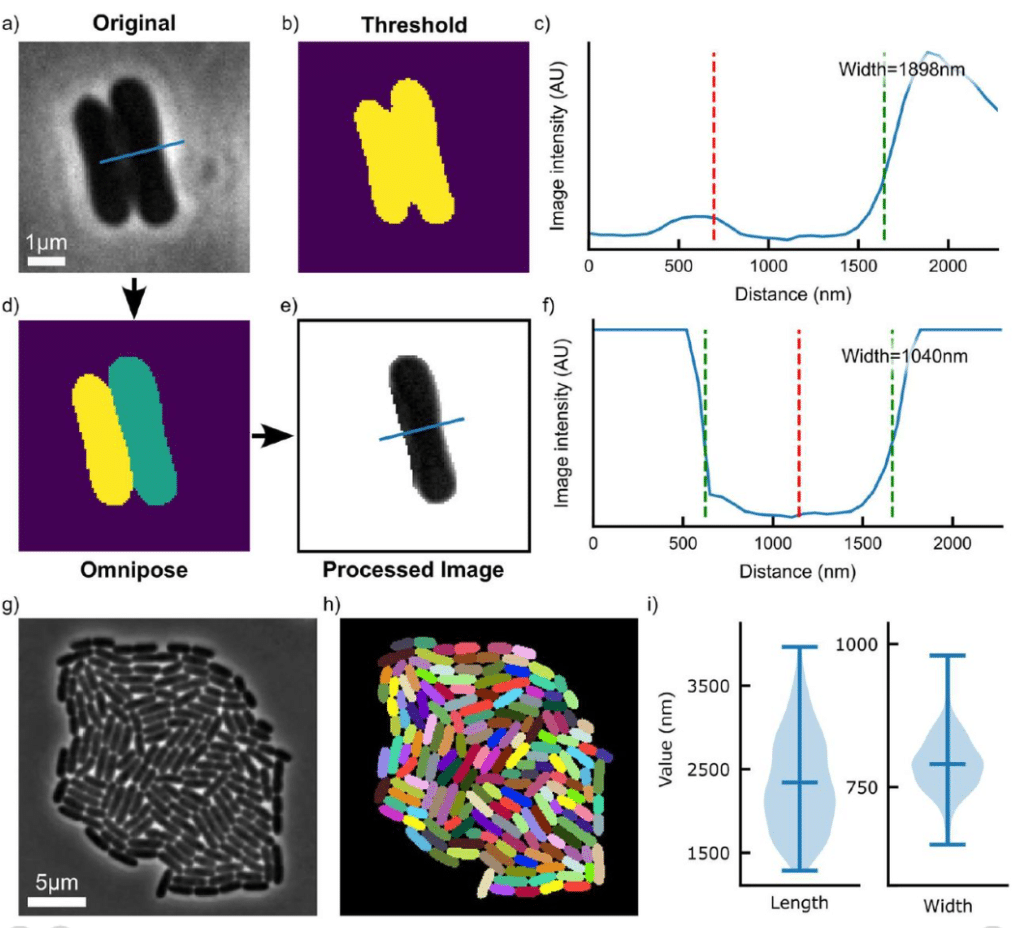
micromorph: a Python toolkit for measurement of microbial morphology
Simone Coppola, Séamus Holden
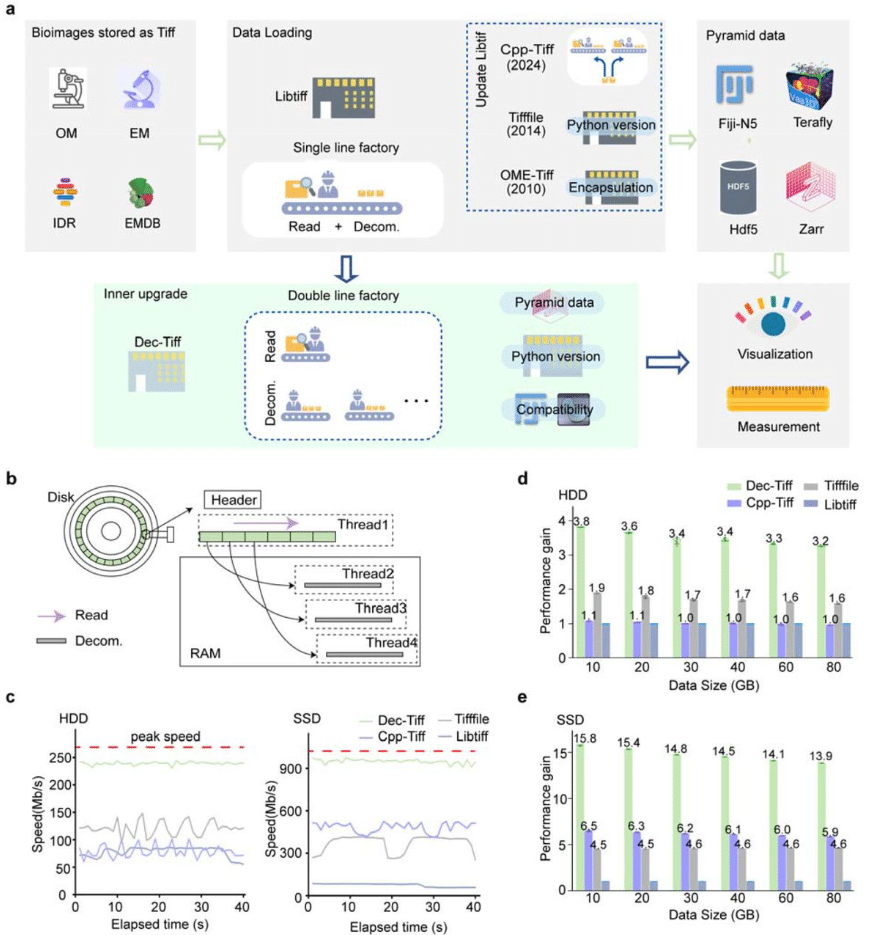
Fast Loading and Transformation of Large-Volume Bioimaging Data Consistently Approaching the I/O Peak Speed Lin Cai, Xuzhong Qu, Hang Zhou, Ning Li, Xianyu Gou, Shiwei Li, Jian Huang, Shasha Guo, Xiuli Liu, Xiaohua Lv, Tingwei Quan, Shaoqun Zeng
A semi-automated pipeline for morphological analysis of myonuclei along single muscle fibers
Esben T. Schroeder, Helia G. Megowan, Madeline Luu, Adam Shuaib, Adam C. Fries, Jake Searcy, Hans C. Dreyer
Quantitative Network Classification with CLaSSiNet Reveals Nanoscale Organizational Principles of the Membrane Skeleton
Yuan Tao, Ruobo Zhou
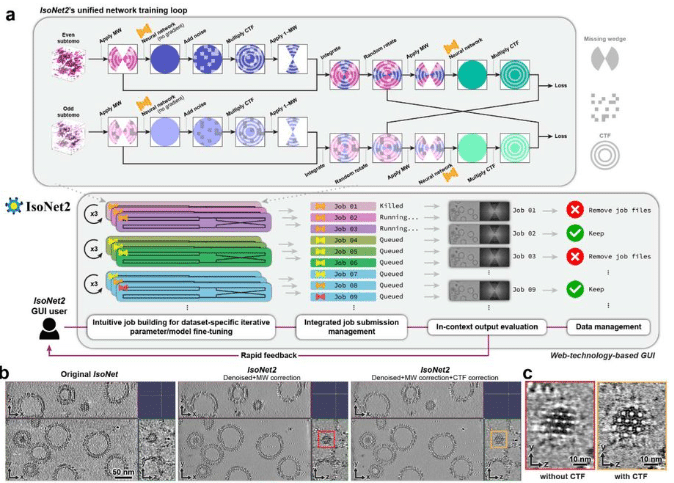
IsoNet2 determines cellular structures at submolecular resolution without averaging
Yun-Tao Liu, Hongcheng Fan, Jonathan Jih, Liam Tran, Xiaoying Zhang, Z. Hong Zhou
Classifying Calcium Imaging Dynamics with Deep Learning: Multi-Frequency Analysis through Quantile-Based Time-Series Network Representations
Caroline L. Alves, Simone Hufgard, Margot Mayer, Helena Dasch, Andriana S. L. O. Campanharo, Loriz Francisco Sallum, Francisco A. Rodrigues, Christiane Thielemann
SIRP: A Self-Supervised Fluorescence Denoising method with Implicit Representation Priors
Yuze Li, Jun Zhu, Pengcheng Xu, Min Guo, Yue Li, Huafeng Liu
Dataset creation for supervised deep learning-based analysis of microscopic images — review of important considerations and recommendations
Christof A. Bertram, Viktoria Weiss, Jonas Ammeling, F. Maria Schabel, Taryn A. Donovan, Frauke Wilm, Christian Marzahl, Katharina Breininger, Marc Aubreville
Rewards-based image analysis in microscopy
Kamyar Barakati, Yu Liu, Utkarsh Pratiush, Boris N. Slautin, Sergei V. Kalinin
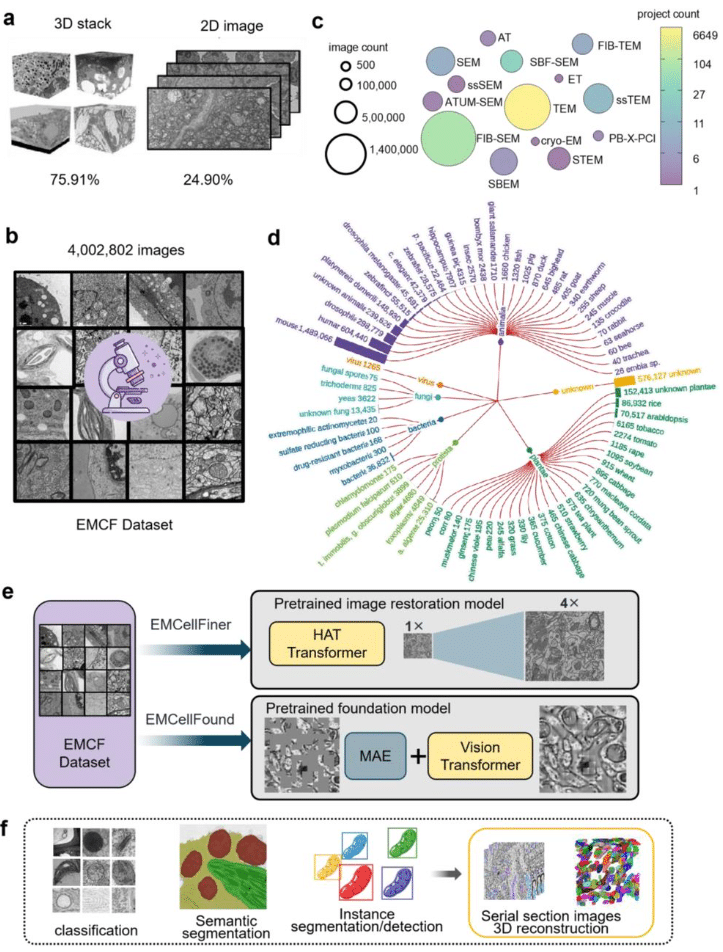
EMCF ecosystem: Towards pretrained foundation model for electron microscopy image analysis
Zeyu Yu, Jiansheng Guo, Feng Liu, Mengze Du, Shan Xu, Guowei Zhang, Li Xie, Bo Han, Zhonghua Chen, Gaoliang Deng, Chen Rui, Yong He, Xuping Feng
IDR searcher: a search engine solution for public image resource
Khaled Mohamed, William Moore, Dominik Lindner, Josh Moore, Jason R. Swedlow, Petr Walczysko, Frances Wong, Jean-Marie Burel
Image-guided alignment of consecutive multi-modal tissue slides
Benedetta Manzato, Claudio Novella Rausell, Gangqi Wang, Nina Ogrinc, Rosalie G.J. Rietjens, Marleen E. Jacobs, Christos Botos, Sebastien J. Dumas, Ton J. Rabelink, Ahmed Mahfouz
MAGNET: an all-in-one foundation model for cross-modal and cross-dimensional microscopic image restoration
Yifan Ma, Tianfeng Zhou, Lanxin Zhu, Chengqiang Yi, Yunshi Zhou, Binbing Liu, Peng Fei
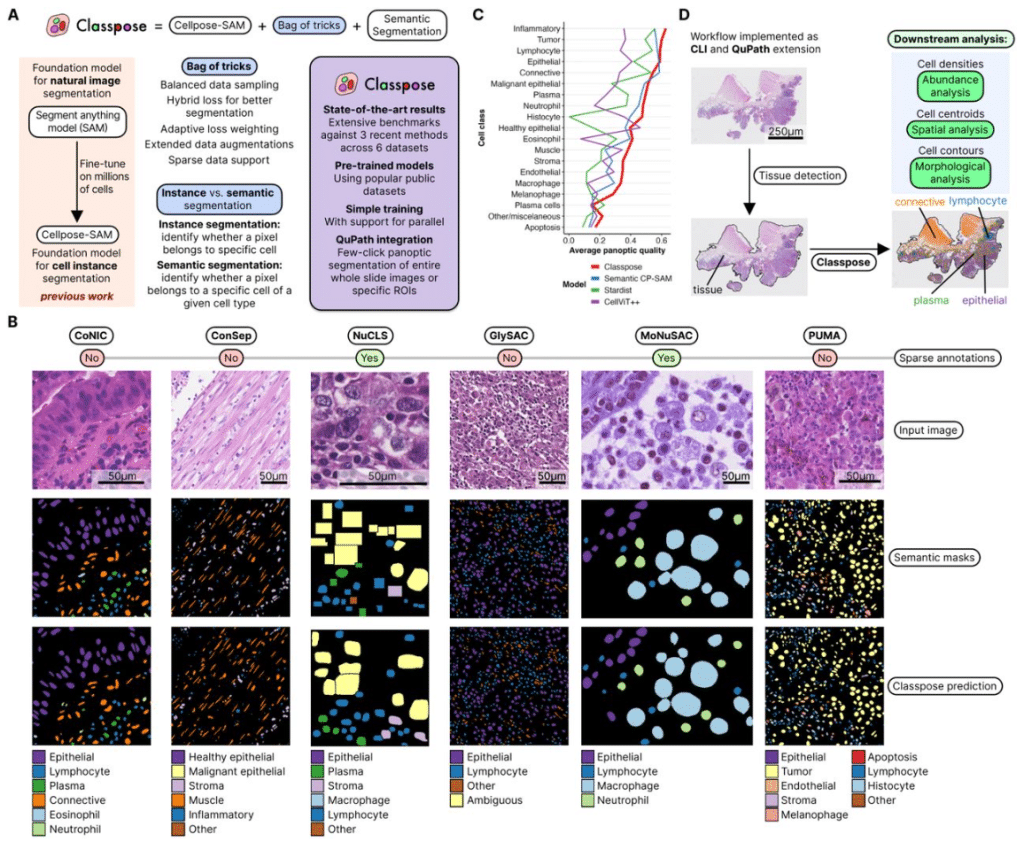
Classpose: foundation model-driven whole slide image-scale cell phenotyping in H&E
Soham Mandal, José Guilherme de Almeida, Nickolas Papanikolaou, Trevor A Graham
An Accessible Python Framework for Real-Time Magnetic Tweezers Microscope Control and Image Processing
James A. London, Abhishek K. Singh, Teague C. Svendsen, Naciye Esma Tirtom, Zachary A. Root, Richard Fishel
Supervised Deep Learning for Efficient Cryo-EM Image Alignment in Drug Discovery with cryoPARES
Ruben Sanchez-Garcia, Alex Berndt, Amir Apelbaum, Judith Reeks, Pamela A Williams, Carl Poelking, Charlotte M Deane, Michael Saur
Leveraging CryoEM and AI-Driven Morphological Feature Analysis for Insights on Bacterial Structures
Sita Sirisha Madugula, Lynnicia N. Massenburg, Spenser R. Brown, Amber N. Bible, Chanda R. Harris, Lance X. Zhang, Kiara Parker, Scott T. Retterer, Jennifer L. Morrell-Falvey, Rama K. Vasudevan, Alexis N. Williams
LivecellX: Corrective Deep Learning for Object-Oriented Single-Cell Analysis in Live-Cell Imaging
Ke Ni, Gaohan Yu, Zhiqian Zheng, Yong Lu, Sophia Hu, Dante Poe, Sijie Zhang, Mia Sanborn, Mostofa Uddin, Zilin Wang, Shiman Zhou, Yanshuo Chen, Xueying Zhan, Weikang Wang, Jianhua Xing
Robust Deep Learning-based 3D Segmentation and Morphological Analysis of Mitochondria using Soft X-ray Tomography
Arun Yadav, Anshu Singh, Aneesh Deshmukh, Pushkar Bharadwaj, Anuj Baliyan, Kate White, Jitin Singla
TransiScope: An Interactive Open-Source Platform for Automated Detection and Analysis of Transient Events in Time-Lapse Microscopy
Rinki Dasgupta, Kaushik Das
CryoSiam: self-supervised representation learning for automated analysis of cryo-electron tomograms
Frosina Stojanovska, Ricardo M. Sanchez, Rasmus K. Jensen, Julia Mahamid, Anna Kreshuk, Judith B. Zaugg
Micro𝕊plit: Semantic Unmixing of Fluorescent Microscopy Data
Ashesh Ashesh, Federico Carrara, Igor Zubarev, Vera Galinova, Melisande Croft, Melissa Pezzotti, Daozheng Gong, Francesca Casagrande, Elisa Colombo, Stefania Giussani, Elena Restelli, Eugenia Cammarota, Juan Manuel Battagliotti, Nikolai Klena, Moises Di Sante, Raghabendra Adhikari, Daniel Feliciano, Gaia Pigino, Elena Taverna, Oliver Harschnitz, Nicola Maghelli, Norbert Scherer, Damian Edward Dalle Nogare, Joran Deschamps, Francesco Pasqualini, Florian Jug
Mastodon: the Command Center for Large-Scale Lineage-Tracing Microscopy Datasets
Johannes Girstmair, Tobias Pietzsch, Vladimir Ulman, Stefan Hahmann, Matthias Arzt, Mette Handberg-Thorsager, Samuel Pantze, Ko Sugawara, Robert Haase, Jean-Yves Tinevez, Pavel Tomancak
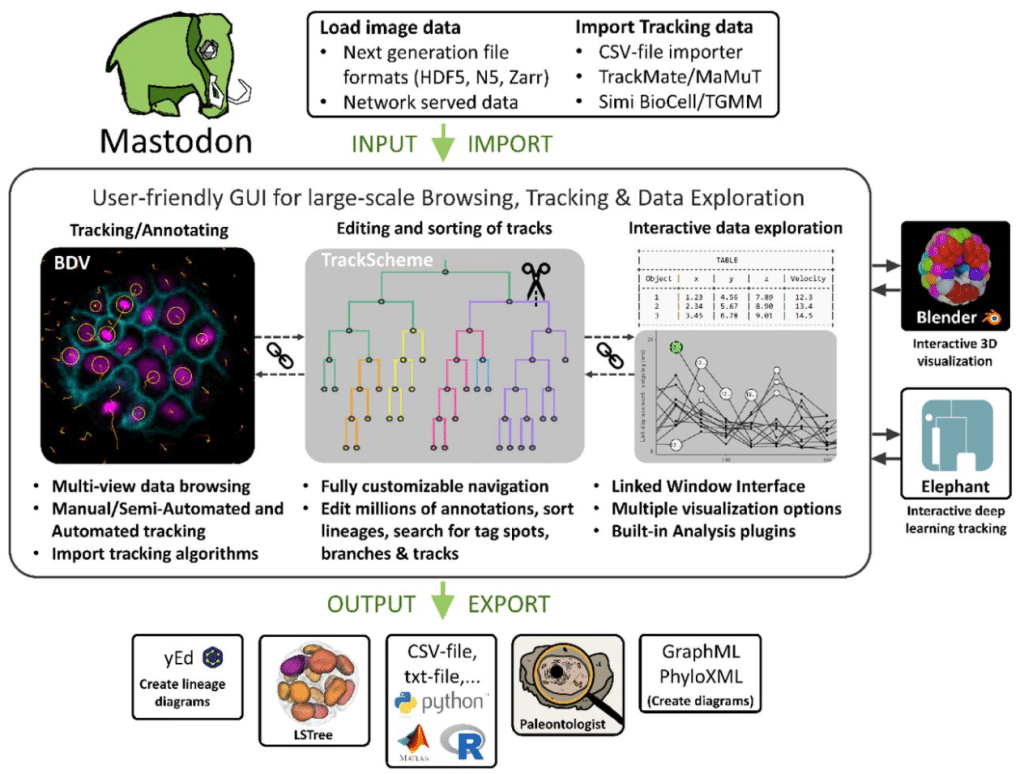
Chart Builder: An Interactive Tool for User Driven Data Visualization in the Electron Microscopy Data Bank
Neli Fonseca, Amudha Kumari Duraisamy, Zhe Wang, Sriram Somasundharam, Minoosadat Tayebinia, Lucas C. de Oliveira, Miao Ma, Jack Turner, Ardan Patwardhan, Gerard J. Kleywegt, Matthew Hartley, Kyle L. Morris
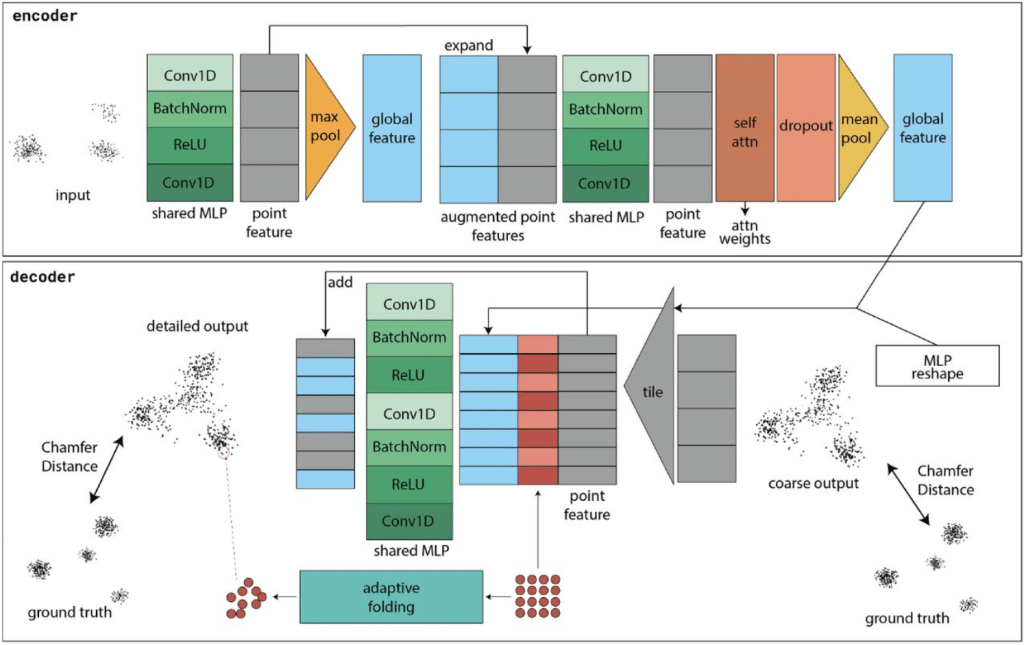
Towards improved particle averaging for single-molecule localization microscopy using geometric deep learning
Diana Mindroc-Filimon, Dominic Helmerich, Patrick Salome, Markus Sauer, Philip Kollmannsberger
FLIM Playground: An interactive, end-to-end graphical user interface for analyzing single cells with fluorescence lifetime imaging microscopy
Wenxuan Zhao, Kayvan Samimi, Melissa C. Skala, Rupsa Datta
MitoEM 2.0: A Benchmark for Challenging 3D Mitochondria Instance Segmentation from EM Images
Peng Liu, Boyu Shen, Liyuan Liu, Qiong Wang, Shulin Zhang, Abhishek Bhardwaj, Ignacio Arganda-Carreras, Kedar Narayan, Donglai Wei


 (No Ratings Yet)
(No Ratings Yet)
