Microscopy preprints: bioimage analysis
Posted by FocalPlane, on 6 February 2026
Here is a curated selection of preprints published (or updated) recently. In this post we focus specifically on bioimage analysis and data management.
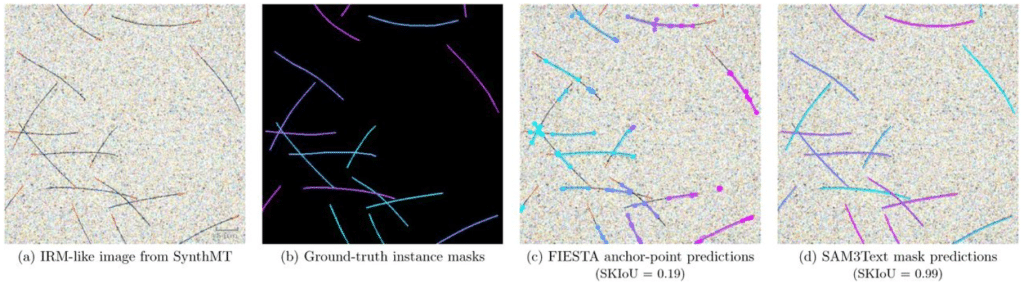
Synthetic data enables human-grade microtubule analysis with foundation models for segmentation
Mario Koddenbrock, Justus Westerhoff, Dominik Fachet, Simone Reber, Felix Gers, Erik Rodner
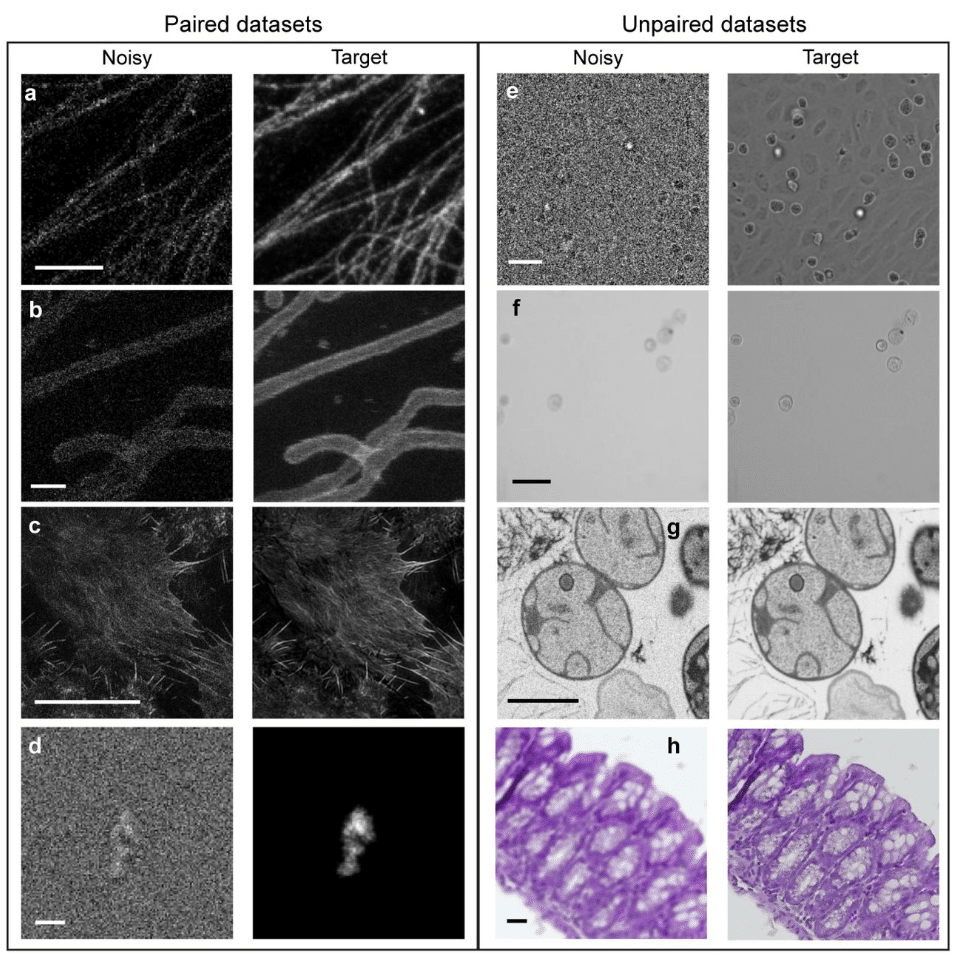
Comparison of Deep Learning Approaches for Extreme Low-SNR Image Restoration
Nasreen Elizabeth Buhn, Sriya Reddy Adunur, Joseph Hamilton, Summer Levis, Guy M. Hagen, Jonathan D. Ventura
HoloBio: A Holographic Microscopy Tool for Quantitative Biological Analysis
Waira Mona, Maria J. Gil-Herrera, Emanuel Mazo, Daniel Córdoba, Sofia Obando, Maria J. Lopera, Rene Restrepo, Carlos Trujillo, Ana Doblas, Raul Castaneda
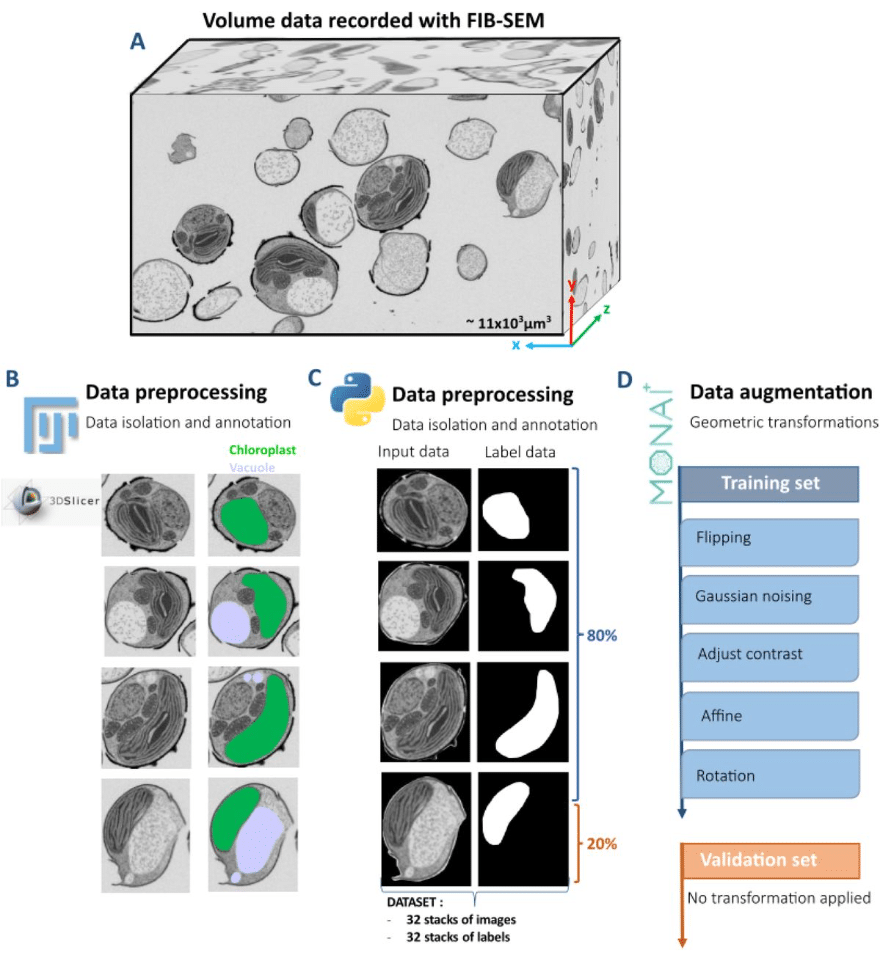
Light Microscopy-Based Organelle Quantification: A Comprehensive Protocol
Suraj Thapilyal, Namdar Hari Kalpana, McMillan Ronald, Jermiah Afolabi, Andrea Marshall, Prasanna Venkhatesh, Ravi Kumar Pujala, Antentor O Hinton Jr, Hailey Parry, Brian Glancy, Prasanna Katti
MorphoLearn: A morphology-driven workflow to decipher 3D electron microscopy segmentation in diatoms
Clarisse Uwizeye, Serena Flori, Jhoanell Angulo, Pierre-Henri Jouneau, Benoit Gallet, Pascal Albanese, Giovanni Finazzi
Smartphone image capture system and image analysis pipelines enable accurate and efficient phenotyping of spaced plant mapping populations
Stella Woeltjen, Molly Hanlon, Keely Brown, Haley Schuhl, Ivan Baxter, Allison Miller
Analysis of 3D epithelial tissue packing reveals correlated patterns of cell height and skewing
Guillaume Pernollet, Quentin Vagne, Juan Manuel García-Arcos, Claire A. Dessalles, Guillaume Salbreux, Aurélien Roux
AutoMorphoTrack: A modular framework for quantitative analysis of organelle morphology, motility, and interactions at single-cell resolution
Armin Bayati, Jackson G. Schumacher, Xiqun Chen
The One Click Wonder: a retrained automated segmentation pipeline that enables quantitative and modular analysis of C. elegans embryos
Palmer Carlyn Bassett, Tobias Evan Verheijen, Angelo Luiz Angonezi, Aude Andriollo, Sebastien Herbert, Gregory Roth, Jeffrey A. Chao, Susan E. Mango
Enabling Real-Time Fluctuation-Based Super Resolution Imaging
Miyase Tekpınar, Jelle Komen, Hana Valenta, Ran Huo, Klarinda de Zwaan, Peter Dedecker, Nergis Tomen, Kristin Grußmayer
A plug-and-play ROI imaging module and deep-learning denoising framework extend three-photon microscopy to 1.7 mm depth
Jixiong Su, Shoupei Liu, Shasha Yang, Yanfeng Zhu, Xinyang Gu, Yaoguang Zhao, Chengyu Li, Min Zhang, Antao Chen, Huiyun Yu, Bo Li
A Self-Supervised Foundation Model for Robust and Generalizable Representation Learning in STED Microscopy
Anthony Bilodeau, Frédéric Beaupré, Julia Chabbert, Kamylle Thériault, Andréanne Deschênes, Jean-Michel Bellavance, Koraly Lessard, Renaud Bernatchez, Paul De Koninck, Christian Gagné, Flavie Lavoie-Cardinal
Enhancing volumetric microscopy with blind computational correction of spatially variant aberrations using neural field representation
Linh Hoang, Zhongqiang Li, Dominique Meyer, Xiankun Lu, Ji Yi
BlueNuclei: automated identification and classification of live and dead transfected neurons using interpretable features
Zhan Zha, Jing Jin, Russell L. Margolis, Daniel Taliun
StomaQuant: Deep Learning-Based Quantification for Stomatal Trait Assessment
Kenny J.X. Lau, Cheng-Yen Chen, Sahanna Muruganantham, Vayutha Muralishankar, Naweed I. Naqvi
Sixteen isotropic 3D fluorescence live imaging datasets of Tribolium castaneum gastrulation
Franziska Krämer, Stefan Münster, Frederic Strobl
A novel attention mechanism for noise-adaptive and robust segmentation of microtubules in microscopy images
Achraf Ait Laydi, Louis Cueff, Mewen Crespo, Yousef El Mourabit, Hélène Bouvrais
AnnotateAnyCell: Open-Source AI Framework for Efficient Annotation in Digital Pathology
Shourya Verma, Aditya Malusare, Mengbo Wang, Luopin Wang, Arpan Mahapatra, Abigail English, Abigail Cox, Meaghan Broman, Simone De Brot, Grant N. Burcham, Deborah Knapp, Deepika Dhawan, Mario Sola, Vaneet Aggarwal, Ananth Grama, Nadia Atallah Lanman
Deep learning-based volumetric denoising enables efficient acquisition of volume electron microscopy
Bohao Chen, Fangfang Wang, Haoyu Wang, Yanchao Zhang, Zhuangzhuang Zhao, Haoran Chen, Hua Han, Xi Chen, Yunfeng Hua
High-fidelity bioimage restoration via adversarial learning
Guillermo Rey-Paniagua, Dariusz Lachowski, Arrate Muñoz-Barrutia
Label-free quantitative phenotyping of hepatic stellate cell activation using holotomography with AI-enabled subcellular segmentation
Sin-hyoung Hong, Junhyung Park, Haesoo Kim, Hyunyoong Moon, Keehang Lee, Hana Lee, Sumin Lee, YongKeun Park
An Open-Source Code To Analyze Mitochondrial Intracellular Distribution From Fluorescence Microscopy Images
Giovanna C. Cavalcante, Alicia J. Kowaltowski
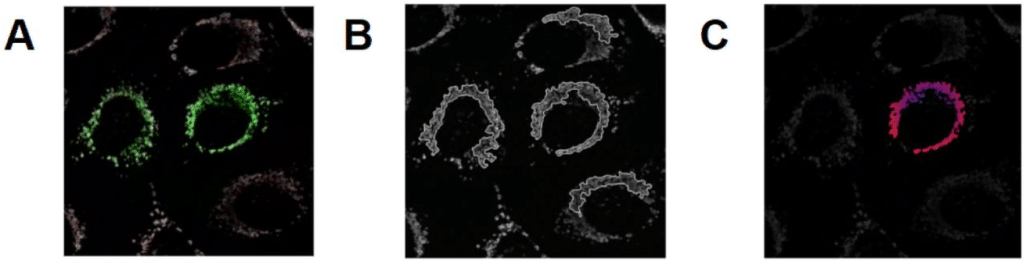
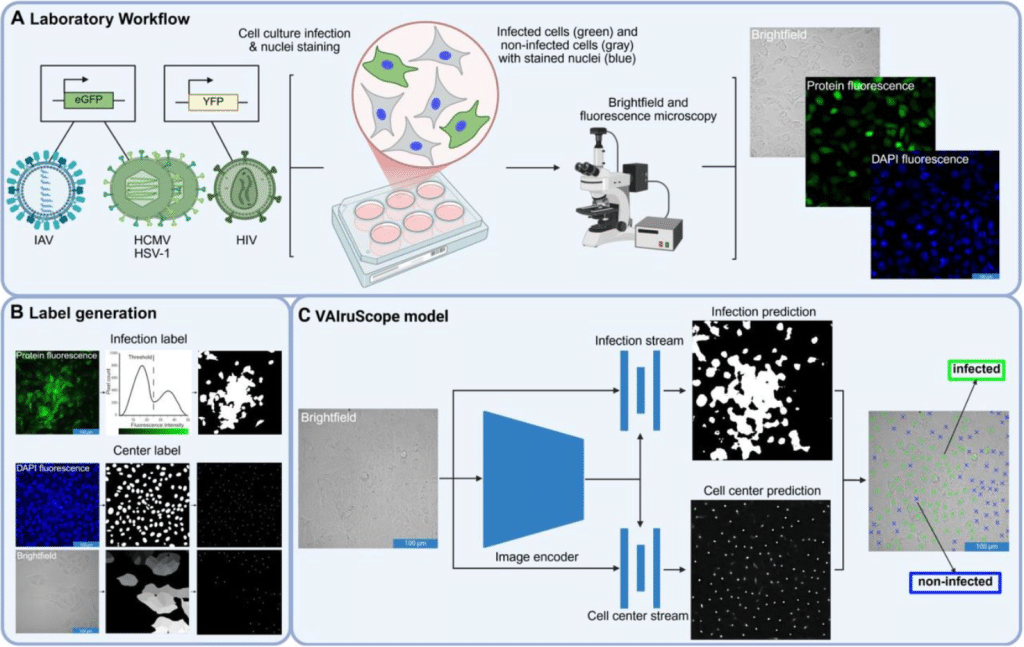
Label-free detection of individual virus-infected cells using deep learning
Juliane Pfeil, Corinna Siegmund, Eva Müller, Shakhnaz Akhmedova, Alexandra Löwe, Anne Kauter, Tobias Tertel, Bernd Giebel, Michael Laue, Vu Thuy Khanh Le-Trilling, Christian Sieben, Mirko Trilling, Roland Schwarzer, Nils Körber
SpatialDINO: A Self-Supervised 3D Vision Transformer that enables Segmentation and Tracking in Crowded Cellular Environments
Alex Lavaee, Arkash Jain, Gustavo Scanavachi, Jose Inacio Costa-Filho, Adam Ingemansson, Tom Kirchhausen


 (No Ratings Yet)
(No Ratings Yet)