Microgametogenesis in four dimensions live-cell imaging of this highly dynamic process in P. falciparum
Posted by FocalPlane, on 27 October 2021
Here is the preLight post from our first preLights – FocalPlane social writing event.
4D live-cell imaging of microgametogenesis in the human malaria parasite Plasmodium falciparum. Sabrina Yahiya, Sarah Jordan, Holly X. Smith, David C. A. Gaboriau, Mufuliat T. Famodimu, Farah A. Dahalan, Alisje Churchyard, George W. Ashdown, Jack Baum
Preprint posted on July 29, 2021 https://www.biorxiv.org/content/10.1101/2021.07.28.454129v1
Background/Introduction
Malaria is a tropical disease caused by parasites of the Plasmodium genus and transmitted by mosquitoes. P. falciparum causes the most dangerous form and accounts for the overwhelming majority of deaths. Mammalian hosts have Plasmodium gametocytes in their bloodstream, which are erythrocytes that are infected with sexual forms of the parasite (male and female). When an infected person is bitten by a mosquito, the mosquito concomitantly ingests the gametocytes with its blood meal. These gametocytes are non-replicative and quiescent in humans, but when they are taken up by mosquitos, they ‘come to life’, transforming into microgametes.
Male Plasmodium microgametogenesis is a fascinating, albeit poorly understood process. The parasite senses its uptake by a mosquito, divides its DNA three times – from one male gametocyte into 8 long and motile gametes – and ejects each gamete via a process termed exflagellation. This is all accomplished in under 20 minutes. The male and female gamete then merge, giving rise to a diploid parasite zygote. Finally, the zygote matures, which is critical for the propagation of the disease into the next human host.
One of the major limitations of understanding this differentiation process is its spatial and temporal scale: gametocytes are very small, and the events happen rapidly. Until this point, information about microgametogenesis has mostly been derived from studies using immunofluorescence and electron microscopy, in which gametocytes were fixed at different time-points after activation. In this preprint, Yahiya and colleagues describe a live 4D microscopy method to observe microgametogenesis in P. falciparum. This imaging pipeline can be used to better study this dynamic process, help find proteins and structures to be targeted by anti-malarial therapies and elucidate the mechanism of action of existing drugs.
Key points of the paper/Main Findings
The authors first developed a 3D+t (4D) imaging pipeline using a widefield microscope to observe microgametogenesis of P. falciparum gametocytes cultured with erythrocytes in real-time and in three dimensions. Tubulin, DNA and the host erythrocyte membrane were labelled with SiR-tubulin (silicon-rhodamine derivative), Vybrant™ DyeCycle™ Vibrant and wheat germ agglutinin, respectively. During image acquisition, where morphology was changing nearly every minute, the microscope was switched back and forth between fluorescence and brightfield illumination to minimize phototoxicity.
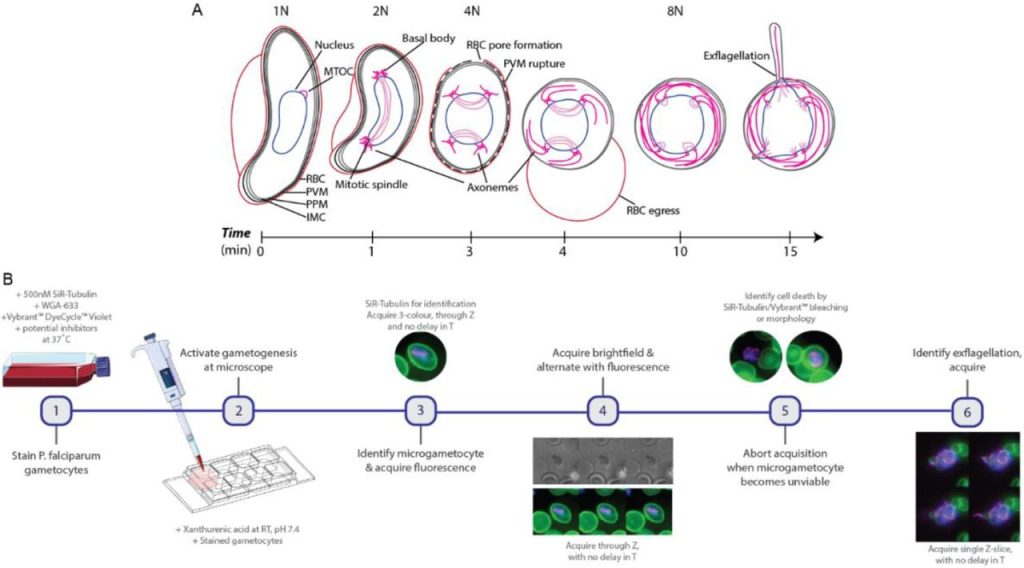
Using this workflow, the authors observed that tubulin polymerized into microtubules during the early stages of microgametogenesis, which also correlated with microgametocyte rounding. Through quantification of DNA using volumetric data, they were able to see that the amount of DNA increased with each nuclear division. In addition, this imaging method allowed them to make the novel observation that segregated DNA was positioned perpendicularly to the basal body tetrads. Tubulin perforated the erythrocyte membrane to form a pore at the spindle pole for microgamete egress. During exflagellation the microgametes were highly motile, leading to data acquisition in 2D rather than 3D time-lapse. However, this process could still be closely observed by visualizing the SiR-tubulin stained axonemes and using brightfield illumination.
To further explore the amenability of their combination of 3D+t (4D) and 2D+t imaging workflow, the authors utilized known anti-malarial drugs to assess their mechanisms of action during microgametogenesis: compounds 1294 and ML 10, inhibitors of Ca2+-dependent protein kinase 4 and cyclic-GMP dependent protein kinase G, respectively. Both compounds were shown to block tubulin polymerization, DNA replication, and microgametocyte rounding, with diverse egress phenotypes.
The authors next explored the role of the proteasome in P. falciparum microgametogenesis using bortezomib, a known proteasome inhibitor active against the asexual replication of Plasmodium. Treatment with bortezomib prevented full cytoskeletal rearrangement, but less completely than with the other two compounds, while also inhibiting DNA replication, microgametocyte rounding, and with mixed effects on success of egress. The microgametes were truncated and shown to only form a maximum of three axonemes. With their imaging method, the authors were able to show the cellular role of the proteasome during microgametogenesis for the first time.
Why is this paper interesting?
This paper will be of interest to malarial researchers as it offers a new combination of 3D+t (4D) and 2D+t imaging and analysis pipeline. This high-resolution live-cell imaging approach gives some new insight into the process of microgametogenesis, including the perforation/pore formation of the host erythrocyte at spindle poles, and the segregation of DNA perpendicular to the basal bodies. In addition, microgametogenesis is a fast process, meaning the parasitology community needs new techniques such as this for visualization. Finally, targeting the proteasome as an antimalarial therapy is relatively novel and may now be desirable to look at in future studies.
Questions to authors
Based on SiR-tubulin staining, it appears as though spindle formation occurs before rounding of the cell. Can the authors speculate on whether tubulin polymerization is causing the rounding of cells?
Can the authors comment on the significance of the spindle being perpendicular to the DNA? Is the spindle dividing the nucleus into two sections?
Inhibition of CDPK4 seemed to inhibit all processes of microgametogenesis; was this all happening at the same time?
Was there a person constantly at the microscope, or was data acquisition automated? Can this pipeline be used or developed for high through-put drug discovery?
Wider context
Looking into the future, it will be of interest to see if this method can be used to study other dynamic developmental processes of other cell types.
Author’s response
Based on SiR-tubulin staining, it appears as though spindle formation occurs before rounding of the cell. Can the authors speculate on whether tubulin polymerization is causing the rounding of cells?
Interestingly, we observed spindle assembly to occur at varied stages of microgametogenesis, both in falciform and rounded parasites. Whilst there is a wealth of data on the shape-shifting nature of gametocytes during their maturation to stage V (Dixon & Tilley, 2021), little is known of the rounding-up process during gametogenesis. The rounding of stage IV gametocyte pointed tips to the rounded tips of the characteristic falciform stage V gametocytes involves microtubule disassembly, in what is likely to be a huge depolymerisation event (Dixon & Tilley, 2021). We speculate something similar may be occurring to enable further rounding up upon activation of gametogenesis, though this certainly requires further study.
Can the authors comment on the significance of the spindle being perpendicular to the DNA? Is the spindle dividing the nucleus into two sections?
The mitotic spindle formed during microgametogenesis spans the length of the parasite cell body, spacing the basal body tetrads, which we observed here with SiR-tubulin. It is well established that the basal bodies lay in close association with the parasite nucleus to enable microgametes to carry a haploid genome along with a developed axoneme through the parasite surface, but little is known of the DNA segregation process. The perpendicular segregation could indeed point to a role of the mitotic spindle dividing segregated DNA and this is a hugely interesting hypothesis that warrants further investigation. With further DNA replication, segregation became increasingly challenging to monitor and it would be fascinating to see if further mitotic spindle assembly enabled simultaneous segregation.
Inhibition of CDPK4 seemed to inhibit all processes of microgametogenesis; was this all happening at the same time?
Being so remarkably complex in nature, microgametogenesis is tightly regulated with a signalling cascade and this includes CDPK4 activity (Fang et al., 2017). Labelled as the ‘master regulator’ of microgamete formation, CDPK4 plays a role in mitotic spindle assembly, initiation of the first genome replication and microgamete motility, acting within only 20 seconds post-activation (Fang et al., 2017). It is therefore no surprise that inhibition of CDPK4 halted all transformation when treating parasites. By blocking the initial stages of microgametogenesis, CDPK4 inhibition clearly prevented all downstream processes. Crucially, this has also been observed in fixed parasites and demonstrates the reproducibility of fixed-cell protocols using our assay.
Was there a person constantly at the microscope, or was data acquisition automated? Can this pipeline be used or developed for high through-put drug discovery?
For the purpose of this study, all acquisition was performed at the microscope, but we do believe new imaging framework is its applicability to high-throughput drug discovery and are excited at the prospect of this. The advent of automated microscopy has hugely transformed the drug discovery landscape and this microgametogenesis imaging protocol is certainly amenable to those technologies. Crucially, we found that acquisiton of fluorescence every few minutes, between brightfield imaging, prolonged microgametocyte viability. By automating acquisition to occur every few minutes at a reduced LED intensity and at randomly selected coordinates, this framework can absolutely be used for phenotypic characterisation of alternative microgametogenesis-blocking drugs (a must in the battle against malaria!).
References
Dixon, M. W. A., & Tilley, L. (2021). Plasmodium falciparum goes bananas for sex. Mol Biochem Parasitol, 244, 111385. doi:10.1016/j.molbiopara.2021.111385
Fang, H. et al. (2017). Multiple short windows of calcium-dependent protein kinase 4 activity coordinate distinct cell cycle events during Plasmodium gametogenesis. Elife, 6, e26524. doi:10.7554/eLife.26524





 (No Ratings Yet)
(No Ratings Yet)