#GliaMorph – a tool to analyse glial cell morphology in 3D
Posted by Elisabeth Kugler, on 17 May 2022
Contents
- What is #GliaMorph?
- What can you do and analyse with #GliaMorph?
- Links to manuscripts and data
- Contributions and Feedback
- Thanks to all contributors and people providing feedback
- References
What is GliaMorph?
GliaMorph is a toolkit implemented as Fiji Macros to allow modular application to process and analyse Müller glia morphology in the retina.
During development glial cells and neurons must spatially and functionally work together to make precise contacts and build a partnership that is necessary for a healthy functioning central nervous system (CNS). Glial cells physically ensheath blood vessels with their endfeet, creating the semi-permeable glia limitans [1]. Importantly, glia physically connect vessels to neurons [2], modulate neurotransmission, and impact neurogenesis [3], [4]. To achieve this, glia cells have elaborate morphologies with apicobasal polarity. Altered glial morphology is a common pathological feature of most neurological disorders, but changed glial cell projections are also observed in healthy ageing [5].
The retina, a component of the CNS, is formed by of seven essential cell types; six neuronal and the principal glial type, the Müller glia (MG). These cells are highly organised into stereotypic discrete tissue layers that establish neuronal circuits and retinal functionality. MG are considered molecular and functional homologues to astrocytes as play critical roles in neuron support, neurovascular function, and brain-retina-barrier health [6], [7].
MG are among the last retinal cell types to mature, integrating into neuronal circuits when neurons are undergoing robust synaptogenesis [8]. However, molecular factors regulating glial morphogenesis have been challenging to identify and sensitive objective computational morphology analysis is needed to understand the processes and factors that drive glia development, health, and disease.
To achieve unbiased analysis of MG morphology in the vertebrate retina we have developed an analysis workflow called “GliaMorph”, which is a modular image analysis toolkit developed in Fiji to perform image pre-processing, semi-automatic ROI selection, apicobasal texture analysis, glia segmentation, and cell feature quantification. Using transgenic and immunohistochemically labelled MG cells, GliaMorph allows for the first time to gain easy-to-use unbiased data understanding and analysis to characterise MG elaboration during development. We moreover, expand GliaMorph application from transgenics to antibodies, from whole-mount to sections, and from zebrafish to mouse retinae.
Working early on with end-user biologists (studying zebrafish, mouse, and human retinae), one main focus was to keep GliaMorph easy to use and adaptable. This means that even though most steps are directly applicable to any data (i.e. increase data comparability using the “SubregionTool”) others might need optimization (i.e. “segmentationTool” will need adaption to the data examined); but this is easily achievable as all steps are implemented as Macros to decrease the barrier for end-users.
To facilitate the use across the community we presented our work early on at conferences, shared our paper on BioRxiv, recorded screencasts, share example data on zenodo, and all code on Github.
Together, the GliaMorph toolkit enables an in-depth understanding on MG morphology, development, and functionality.
What can you do and analyse with #GliaMorph?
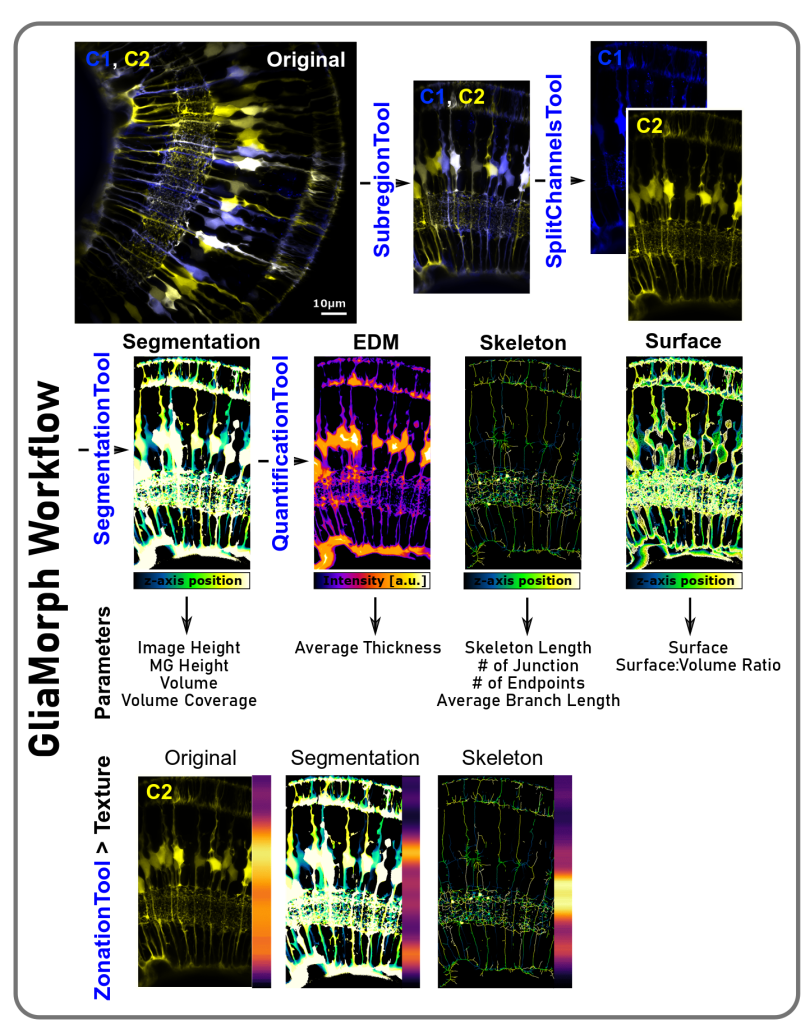
a) Make images more comparable: Working with 3D stacks in biological tissues, direct image comparison is often not possible. Thus, we developed a tool called the “SubregionTool” that allows you to draw a line and then use this line to automatically rotate and 3D-crop your stack, so all images will be comparable between samples and groups. Other steps include things like 90 degree rotation or splitting of multi-channel images.
b) Improve image quality: When working with confocal data, you might want to perform Point-Spread-Function deconvolution prior to improve image quality before subsequent segmentation. To achieve this, we produced the “DeconvolutionTool” that provides a GUI and intuitive input selection for deconvolution, which then uses existing Fiji Plugins namely the “Diffraction PSF 3D” and DeconvolutionLab2 [9].
c) Extract glia cells by image binarization: Following some processing, it is time to extract glia from the image background, which is called segmentation. With #GliaMorph we present some segmentation workflows such as “step7_SegmentationTool” or “EKugler_SegmentationTest”, but as image segmentation is highly dependent on the input data (e.g. transgenic vs immunohistochemistry, whole-mount vs sections, zebrafish vs mouse), segmentation will need to be optimized to your data of interest.
d) Extract parameters to describe overall image parameters: Using segmented data as input images, the next tool called “QuantificationTool” extracts image parameters based on the segmented image, Euclidean distance maps, and the extracted skeleton.
- Image Height (IN) [µm]: Total height of the image
- MG Height (MGN) [µm]: Radial extension of MG
- Number/Count (NN): Number of objects in ROI
- Volume (VN) [µm3]: Volume of object voxel, derived after segmentation
- Volume Coverage (VCN) [%]: Percentage of image volume covered with MG (lowest = 0; highest = 100)
- Surface (SN) [µm3]: Number of object surface voxels, derived after segmentation (given in [µm3] for comparability between experiments)
- Surface:Volume Ratio (S:VN): Ratio of surface to volume (lowest = 0; highest = 1)
- Thickness (TN): Distance from the local centreline to the corresponding surface
- Skeleton length (LN) [µm]: Skeleton voxels (given in [µm] for comparability between experiments)
- # of Junction (JN): Number of points where 2/more sub-branches branch off
- # of Endpoints (EPN): Number of blind-ended object points
- Average Branch Length (BL) [µm]: Average length of individual skeleton branches
e) Analyse apicobasal texture:
We next sought to examine MG apicobasal height by reducing 3D image dimensionality to a one-voxel wise or vector representation by a reduction in x and y with the GliaMorph tool called ZonationTool. This allows visual assessment of apicobasal properties, which are highly insightful, particularly when applied to original, segmented, and skeletonised data.
Links to manuscripts and data:
• BioRxiv manuscript protocol: https://biorxiv.org/cgi/content/short/2022.05.05.490765v1
• Example Data: Data Link: https://zenodo.org/record/5747597 DATA DOI: 10.5281/zenodo.5747597
• Minimum Example Data Link: https://zenodo.org/record/5735442
• YouTube screencasts: https://www.youtube.com/hashtag/gliamorph
(recorded by Sara Beqiri and Karim Nizam, 2022)
Contributions and Feedback
GliaMorph tools are meant to be used and useful, the code and tools are meant to change and be adaptable. Please help us make the most out of #GliaMorph by giving us feedback and testing the tool:
- Raise issues for improvements and create pull requests for code adaptions in Github (as described by Robert Haase: https://focalplane.biologists.com/2021/09/04/collaborative-bio-image-analysis-script-editing-with-git/).
- Contribute to the discussion (https://github.com/ElisabethKugler/GliaMorph/discussions).
- Use the image.sc forum for discussions / questions / how-to’s (https://forum.image.sc/)
- Please use the hashtag #GliaMorph (especially on social media), so we can communicate effectively around the tool.
- For specific questions, please contact kugler.elisabeth[at]gmail.com.
Thanks to all contributors and people providing feedback
Code Author: Elisabeth Kugler
Project Leads: Elisabeth Kugler (code, formal analysis, supervision) and Ryan MacDonald (data, resources, supervision)
Project Contributors: Eva-Maria Breitenbach (tester), Alicia Carrington (data and tester), Isabel Bravo (data and tester), Stefania Marcotti (code: https://github.com/OakesLab/AFT-Alignment_by_Fourier_Transform), Brian M. Stramer (resources), and Pierre Mattar (data and resources).
Feedback: Cyril Eleftheriou (data and tester), Alessandro Felder (code advice), and Robert Haase (code advice).
Contact: kugler.elisabeth[at]gmail.com
Elisabeth Kugler is a Research Fellow at UCL, working at the interface of biology and image analysis.
website | @KuglerElisabeth
Ryan B MacDonald is a principal investigator at the UCL Institute of Ophthalmology interested in beer standing how glial cells support healthy vision.
website| @MacDonald_Lab
References
[1] N. Kutuzov, H. Flyvbjerg, and M. Lauritzen, “Contributions of the glycocalyx, endothelium, and extravascular compartment to the blood–brain barrier,” PNAS, vol. 115, no. 40, pp. E9429–E9438, Oct. 2018, doi: 10.1073/pnas.1802155115.
[2] M. Zonta et al., “Neuron-to-astrocyte signaling is central to the dynamic control of brain microcirculation,” Nature Neuroscience, vol. 6, no. 1, Art. no. 1, Jan. 2003, doi: 10.1038/nn980.
[3] P. Argente-Arizón, S. Guerra-Cantera, L. M. Garcia-Segura, J. Argente, and J. A. Chowen, “Glial cells and energy balance,” J. Mol. Endocrinol., vol. 58, no. 1, pp. R59–R71, 2017, doi: 10.1530/JME-16-0182.
[4] S. Falk and M. Götz, “Glial control of neurogenesis,” Current Opinion in Neurobiology, vol. 47, pp. 188–195, Dec. 2017, doi: 10.1016/j.conb.2017.10.025.
[5] R. R. Martins, M. Zamzam, M. Moosajee, R. Thummel, C. M. Henriques, and R. B. MacDonald, “Müller Glia regenerative potential is maintained throughout life despite neurodegeneration and gliosis in the ageing zebrafish retina,” Oct. 2020. doi: 10.1101/2020.06.28.174821.
[6] E. Newman and A. Reichenbach, “The Müller cell: a functional element of the retina,” Trends Neurosci, vol. 19, no. 8, pp. 307–312, Aug. 1996, doi: 10.1016/0166-2236(96)10040-0.
[7] E. C. Kugler, J. Greenwood, and R. B. MacDonald, “The ‘Neuro-Glial-Vascular’ Unit: The Role of Glia in Neurovascular Unit Formation and Dysfunction,” Frontiers in Cell and Developmental Biology, vol. 9, p. 2641, 2021, doi: 10.3389/fcell.2021.732820.
[8] P. R. Williams et al., “In vivo development of outer retinal synapses in the absence of glial contact,” J Neurosci, vol. 30, no. 36, pp. 11951–11961, Sep. 2010, doi: 10.1523/JNEUROSCI.3391-10.2010.
[9] D. Sage et al., “DeconvolutionLab2: An open-source software for deconvolution microscopy,” Methods, vol. 115, pp. 28–41, 15 2017, doi: 10.1016/j.ymeth.2016.12.015.


 (No Ratings Yet)
(No Ratings Yet)