VollSeg and its extension Napari plugins
Posted by varun kapoor, on 15 January 2023
In this article we present the Napari plugin VollSeg and its ongoing extension plugins that we are creating as a part of the CZI grant awarded successively to the non-profit company KapoorLabs, Paris. VollSeg is a Napari plugin for performing semantic, instance segmentation and denoising using the seed pooling approach that requires StarDist and either a U-Net trained model or a CARE trained denoising model to perform semantic segmentation. This plugin was developed in collaboration between Varun Kapoor (Kapoorlabs, Paris), Jakub Sedzinski (University of Copenhagen, Denmark , Mari Tolonen (University of Copenhagen, Denmark) and Daisuke Inou (Kyushu University, Tokyo) and is funded by CZI, Napari plugin accelerator and foundation grants.
VollSeg segmentation is especially suited for 3D segmentation of membrane/nuclei labelled cells that are irregular in shape. VollSeg can also be used to segment tissues that are imaged using transmitted light, fluorescent or light sheet microscopy.
To find a suitable mode and model for running VollSeg please see our table and based on your use case choose the appropriate model combination to get optimal segmentation results. The advantage of providing the segmentation tool as a plugin is that it allows for several model combinations that can be tested on a subset of the whole dataset prior to applying that chosen combination on the entire imaging data.
Denoising and segmentation is usually the first step towards any data and image analysis pipeline for microscopy data, this step is important as the overall accuracy of the image analysis workflow depends upon the segmentation accuracy attained at this stage. After this stage we are developing specialized plugins that use the segmentation results coming from this plugin and create an automated and interactive workflow for specific use cases. Our current use cases of interest are
- Quantification of dynamic instability parameters of microtubule and actin filaments that are imaged in vitro, such filaments are analyzed using Kymographs to quantify the growth, shrink rates and catastrophe and rescue frequency. Usual practise in the field is to obtain the Kymographs and then manually fit lines to compute these parameters, a task that is manual, tedious, time-consuming and error-prone. In our approach we use VollSeg trained model to segment the kymograph and then use outlier removal technique called RANSAC for fitting linear and quadratic functions on the found inliers to automatically quantify the dynamic instability parameters in batch. This plugin is available from Napari hub and video tutorial on using this plugin can be found on our Youtube channel.
- Tissues or organoids are images using transmitted light microscopy or fluorescent or light sheet microscopy, given our pre-trained models we can segment such tissues in 2D, 3D and 2D+time and 3D+time datasets directly in the plugin. Post segmentation we use the Napari plugin skan for performing the skeletonization of the tissue and track the found skeleton end points to quantify the growth rate of such tissues/organoids. Prior to our work this functionality was only available for 2D+ time datasets as our TrackMate extension, BTrack that was used in this publication.
- Post segmentation tracking is performed using the segmented images for doing the frame to frame linking. TrackMate in Fiji universe is a popular plugin for doing tracking, track analysis and also supports customized extensions written by several developers. We provide a VollSeg extension for performing track analysis using customised reads including statistics and temporal statistics, analysis of brownian and non-brownian motion trajectories, frequency analysis of cell intensity oscillations etc by using the outputs of TrackMate, i.e., the Tracking xml file and the spots, tracks and edges csv file exports that are then imported into our plugin to provide with more track analysis of these trajectories and visualization of the trajectories using Napari tracks layer.
The vollseg-napari-mtrack plugin after installation shows up as shown below, a directory containing kymographs can be dragged and dropped and further analysis can be performed as detailed in this video. To make the analysis simpler please create the kymographs of the plus and minus ends of the microtubules separately.
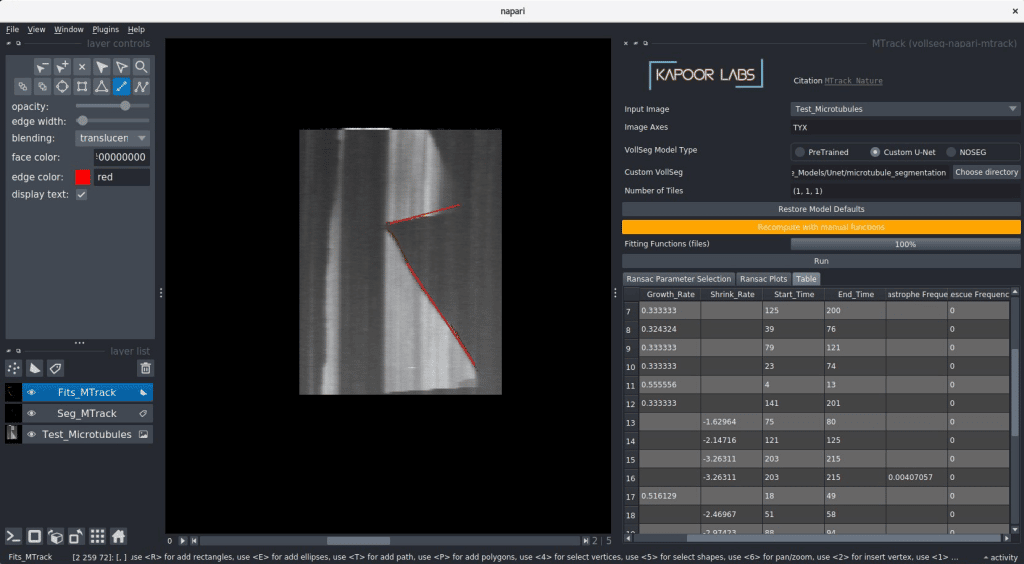
The vollseg-napari-trckmate plugin after installation would look as shown below, for creating only statistical plots you can skip loading the optional Raw and segmentation images and only load the xml and csv files coming out of the TrackMate plugin. A more elaborate video detailing this steps can be found here.
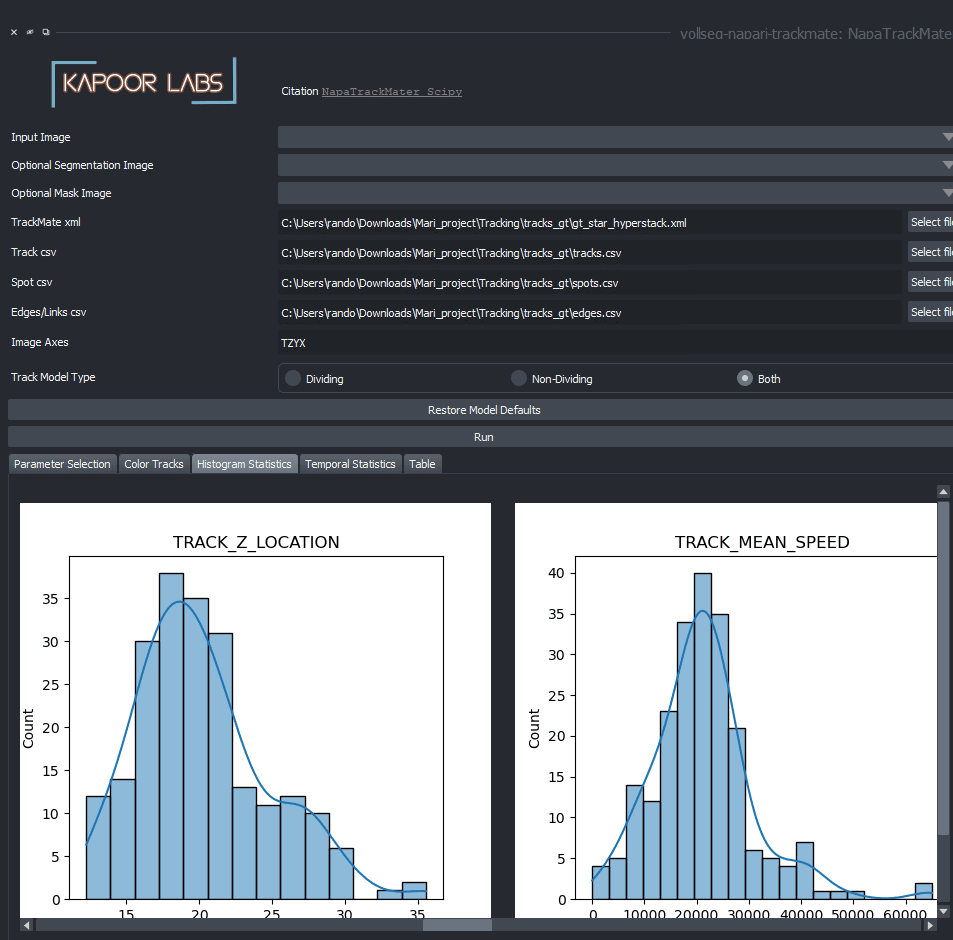
Installation instructions
VollSeg is available as a Napari plugin called vollseg-napari on the Napari plugin hub, to install it open Napari and search for the plugin in plugins drop down menu and click install. We also provide wheels for vollseg-napari, to install vollseg-napari via command line please follow these installation instructions. You can find all the VollSeg related extension plugin on our Napari hub collection page.


 (No Ratings Yet)
(No Ratings Yet)