Fast4DReg – to the rescue of your drifty microscopy data
Posted by Joanna Pylvänäinen, on 10 March 2023
Fast4DReg to the rescue of your 4D microscopy data
In life sciences, researchers use microscopes to study living organisms, such as cells or small animals. These live cell imaging experiments are usually performed over several hours, exposing the experiment to changes in the sample and in the microscope surroundings – causing the data to drift. This drift can be caused, for example, by temperature changes leading to thermal expansion of the microscope mechanical components or by the movement of the sample itself in the field of view. To correct such movement, we developed a tool called Fast4DReg (Pylvänäinen et al., 2023), an open-source drift correction tool available in Fiji (Schindelin et al., 2012). Fast4DReg corrects 4D data by creating intensity projections along multiple axes and estimating the drift between frames using two-dimensional cross-correlations (Figure 1).
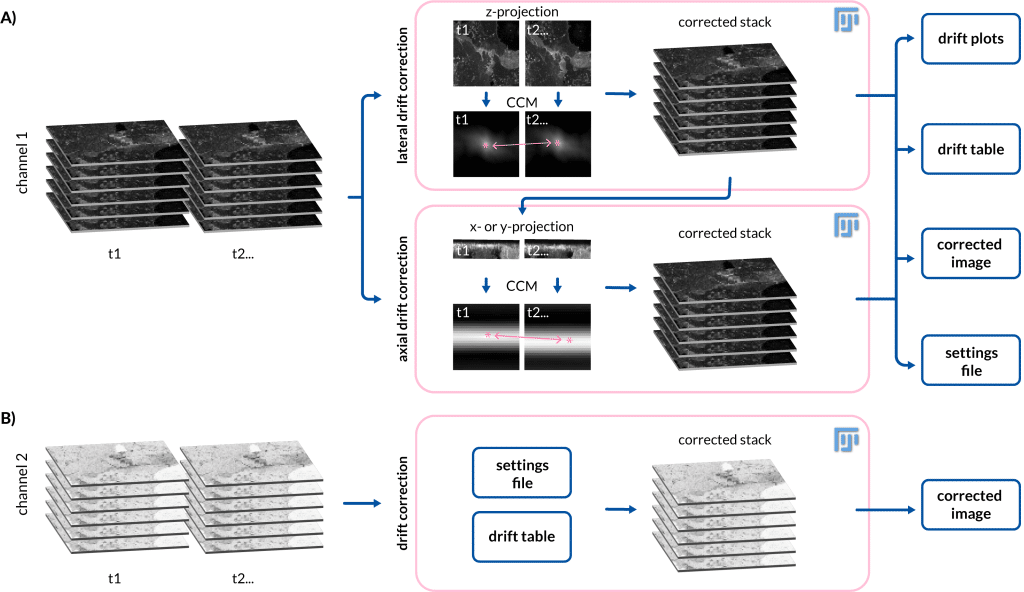
The need for Fast4DReg arose from real issues with our 4D data
The need to start developing Fast4DReg in the first place arose from an actual issue in our lab. My colleagues Gautier Follain and Sujan Ghimire are studying how cancer cells extravasate the vasculature during cancer metastasis. In one of their more complicated experimental setups, they mixed different cell lines, set up a flow system and acquired 3D videos. As every acquired video was several hours long, the flow setup moved while it was imaged causing lateral drift. In addition to the lateral drift, the microscope that was used to image this sample had a stage issue causing oscillations of the stage that led to axial drift of the data. We tried several different tools to correct the drift, but we could not find a tool that worked sufficiently well for our data. Therefore, we needed to build our own solution.
My supervisor Guillaume Jacquemet has been collaborating with Ricardo Henriques for some time now. Henriques lab has published a software package, NanoJ (Laine et al., 2019), that includes a cross-correlation-based drift correction tool for the correction of 2D data. Guillaume had successfully used this tool before for the correction of 2D drift and was thinking that maybe if expanded to support 3D data it could be a solution to our problem. To test this Romain Laine, a postdoc in Henriques’ lab, used one of his lunch breaks (or this is what I was told 😀 ) to test if NanoJ 2D cross-correlation-based drift correction could be extended to support 3D videos. By the end of his lunch break, he had written a few lines, that would allow us to do just that – great! This was when I joined the project. My task was to take these lines of code and expand the to a functional drift correction tool, that would support the correction of drift in multiple datasets in xy- and z-directions in batch as well as the possibility of using already defined drift settings for other datasets. Additionally, we edited the code so that it allows the correction of multichannel data. For a better user experience, I created a user interface, that helps in getting started with Fast4DReg. I value transparent documentation and therefore we spend time making this tool easily findable, accessible, and user-friendly. We published the Fast4DReg in the Journal of Cell Science (Pylvänäinen et al., 2023) where we demonstrated the performance of Fast4DReg, and of course, tell everyone about this new wonderful tool.
Fast4DReg surprised us all with its performance
To test the performance of Fast4DReg, we created a dataset with synthetic drift and different levels of noise. By using this dataset, we showed that Fast4DReg over-performed other tools we tested previously, such as Fijiyama (Fernandez & Moisy, 2021) or Correct 3D Drift (Parslow et al., 2014) (Video 1). One of the main advantages of Fast4DReg is that it measures the amount the drift using 2D projections, and therefore it performs much faster than tools that use 3D volumes for drift estimation.
We also demonstrate that the correction ability of Fast4DReg was not affected by noise until the signal-to-noise ratio (SNR) was below 2. Additionally, by opting for the average projection as the projection type, we successfully corrected videos with an SNR as low as 1.6. This is particularly valuable when dealing with live cell imaging data which frequently encounters low SNR issues resulting from efforts to minimize phototoxicity.
Next, we tested the performance of Fast4DReg on real biological data. My colleague Gautier was imaging cancer cells migrating inside the mouse lung vasculature in a precision-cut lung slice, which was impossible to immobilize completely. This data also had multiple channels (cancer cells and vasculature) and time points. In this dataset, the cancer cells were motile, while the vasculature stayed more or less stable. Therefore we decided to estimate the drift in this dataset using the vasculature as a reference, and the generated drift table was then applied to the channel with cancer cells (Figure 2). This way, we successfully corrected the drift in this dataset (Video 2).
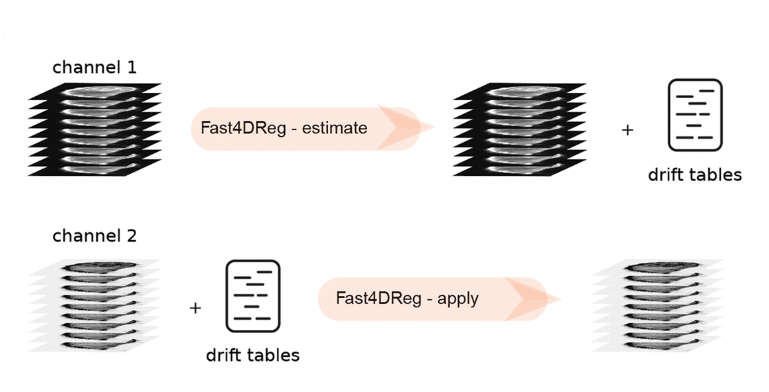
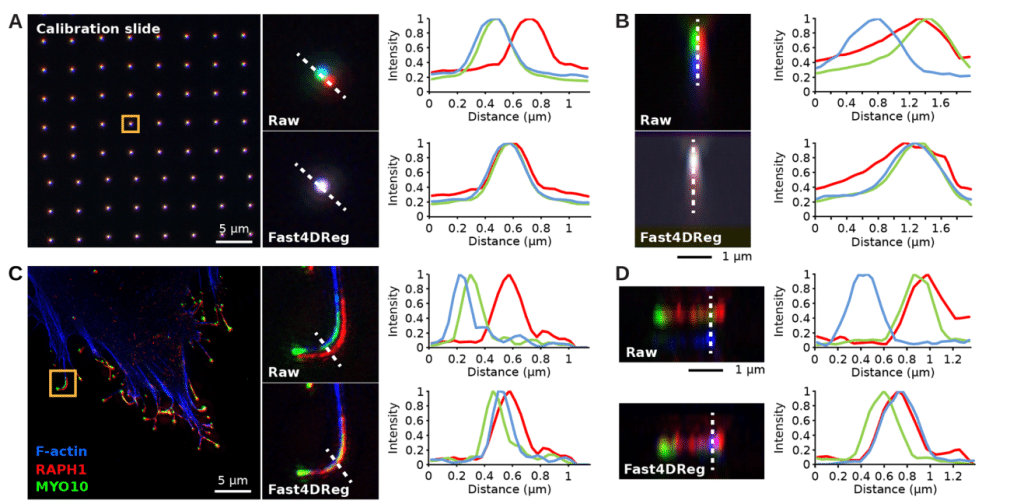
Fast4DReg can also correct misaligned channels
To test how versatile Fast4Dreg is, we tested if it could be used to align 3D multichannel images instead of 3D videos, by converting the channels into time frames prior to correction. First, we used a calibration slide to estimate how much the channels were misaligned. Next, we corrected an image with a similar misalignment using the drift table acquired from correcting the calibration slide image using Fast4DReg. We used line intensity profiles to quantify and show that we were able to align the channels in the calibration slide and apply the correction to a cell image (Figure 3). We developed Fast4DReg with the capability of batch-correcting misaligned images acquired under the same conditions as the calibration slide. Compared to correcting the data frame by frame, this saves massive amounts of time. This two-step channel correction method using a calibration slide may be particularly advantageous for colocalization studies, where accurate channel alignment is crucial for obtaining reliable results.
Software availability
With Fast4DReg, we hope to make the process of multidimensional data registration more straightforward and faster and, therefore, more accessible to the community.
Fast4DReg is available through:
In Github in addition to the most recent version of Fast4DReg, we provide detailed step-by-step instructions on how to use the tool. In Zenodo, we share all data used for testing the tool in our recent publication in the Journal of Cell Science: Fast4DReg – fast registration of 4D microscopy datasets.
Acknowledgments
I want to thank everyone who helped me to get this publication together, Sujan and Gautier for providing data, Romain and Bruno for helping me with the code, and Guillaume and Ricardo for excellent guidance. This was truly a collaborative effort, and I feel grateful to have the opportunity to work with all these wonderful people. Thanks!

posted by
Joanna Pylvänäinen
PhD student at Cell Migration Lab
Åbo Akademi University, Turku, Finland
(picture by Solveig Eriksson)
Literature
Fernandez, R., & Moisy, C. (2021). Fijiyama: A registration tool for 3D multimodal time-lapse imaging. Bioinformatics (Oxford, England), 37(10), 1482–1484. https://doi.org/10.1093/bioinformatics/btaa846
Laine, R. F., Tosheva, K. L., Gustafsson, N., Gray, R. D. M., Almada, P., Albrecht, D., Risa, G. T., Hurtig, F., Lind\aas, A.-C., Baum, B., Mercer, J., Leterrier, C., Pereira, P. M., Culley, S., & Henriques, R. (2019). NanoJ: A high-performance open-source super-resolution microscopy toolbox. Journal of Physics D: Applied Physics, 52(16), 163001. https://doi.org/10.1088/1361-6463/ab0261
Parslow, A., Cardona, A., & Bryson-Richardson, R. J. (2014). Sample drift correction following 4D confocal time-lapse imaging. Journal of Visualized Experiments: JoVE, 86. https://doi.org/10.3791/51086
Pylvänäinen, J. W., Laine, R. F., Saraiva, B. M. S., Ghimire, S., Follain, G., Henriques, R., & Jacquemet, G. (2023). Fast4DReg – fast registration of 4D microscopy datasets. Journal of Cell Science, 136(4), jcs260728. https://doi.org/10.1242/jcs.260728
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J.-Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P., & Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9(7), 676–682. https://doi.org/10.1038/nmeth.2019


 (3 votes, average: 1.00 out of 1)
(3 votes, average: 1.00 out of 1)

I tried this on 4D time-lapse bead data which was used to characterize drift in an upright spinning disk microscope and it worked really well. Of course the SNR was excellent, so no trouble there. The Github doc was excellent: very clear and well written. Kudos to the team who developed this. I will expore it with more complex live cell data in future. Thank you!