Featured image with Simon Desiderio
Posted by FocalPlane, on 21 April 2023
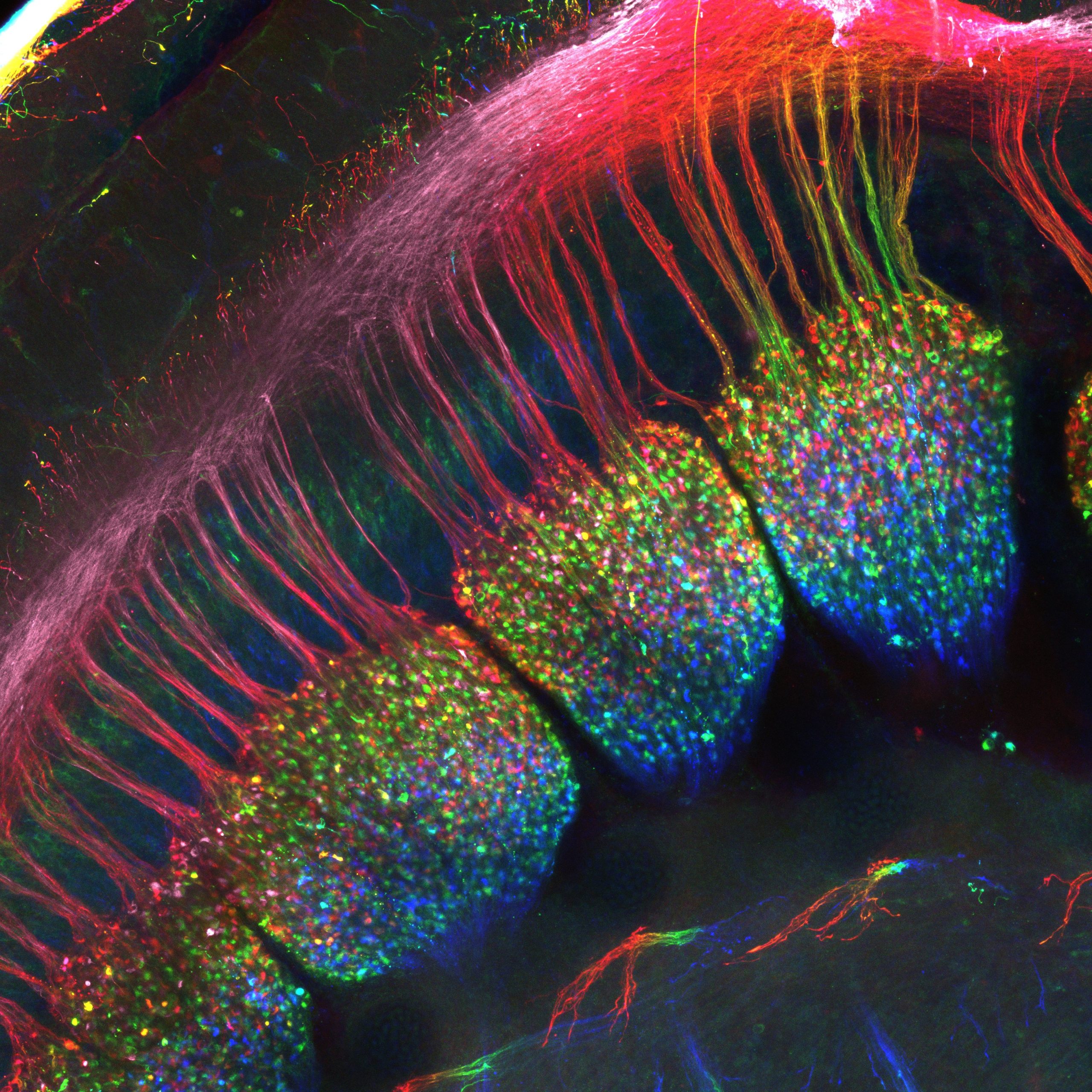
Our featured image shows the developing dorsal root ganglia of an E12.5 mouse embryo from Simon Desiderio. A whole-mount immunostaining was performed with a Peripherin antibody to label sensory neurons within the ganglia as well as their projections to the spinal cord that constitute these “dorsal roots”. The embryo has been cleared using the SeeDB method to allow visualization of the ganglia. The sample was acquired on a LSM710 confocal microscope and this final image represents a maximum intensity projection which has been depth color-coded.
We caught up with Simon Desiderio to find out more about his research career and what he is excited about in microscopy.
Research career so far: I completed my PhD at the University of Brussels, where I investigated the developmental contribution of the transcription factor Prdm12 to the neurogenesis of spinal cord and dorsal root ganglia. I then moved for a postdoc at the Institute for Neurosciences of Montpellier (INM) where I studied the mechanisms of sensory neurogenesis and the development of tactile sensory neurons. I am now back at the University of Brussels for a second postdoc, still trying to understand the molecular mechanisms behind the setup of tactile sensory neurons.
Current research: My current aim in the laboratory of Professor Eric Bellefroid is to identify molecular determinants playing critical functions in the neurogenesis of tactile sensory neurons as well as in the establishment of their unique functional properties. We mainly focus on transcription factors and try to get a clear picture of the developmental features that they modulate.
Favourite imaging technique/microscope: I do not have extensive experience with many microscopes or imaging techniques, but I always enjoy spending hours in the dark with a confocal microscope!
What are you most excited about in microscopy? I am very excited by the possibilities now offered by optical clearing techniques to directly visualise complete or thick biological samples. It offers unprecedent ways to investigate biological questions and to look at parameters that would be hardly measurable on classic cryotome slices. The increasing genetic toolbox allowing selective spatial or temporal labelling of cell populations (AAV, brainbow, etc) as well as the tools and equipment increasing the resolution of immunostainings (airyscan, spaghetti monster fluorescent proteins, etc) make also good promises for the future of microscopy.


 (No Ratings Yet)
(No Ratings Yet)