Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 24 January 2025
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
CellPhePy: a Python implementation of the CellPhe toolkit for automated cell phenotyping from microscopy time-lapse videos
Laura Wiggins, Stuart Lacy, Graeme Park, Joanne Marrison, Ben Powell, Beth Cimini, Peter O’Toole, Julie Wilson, William J. Brackenbury
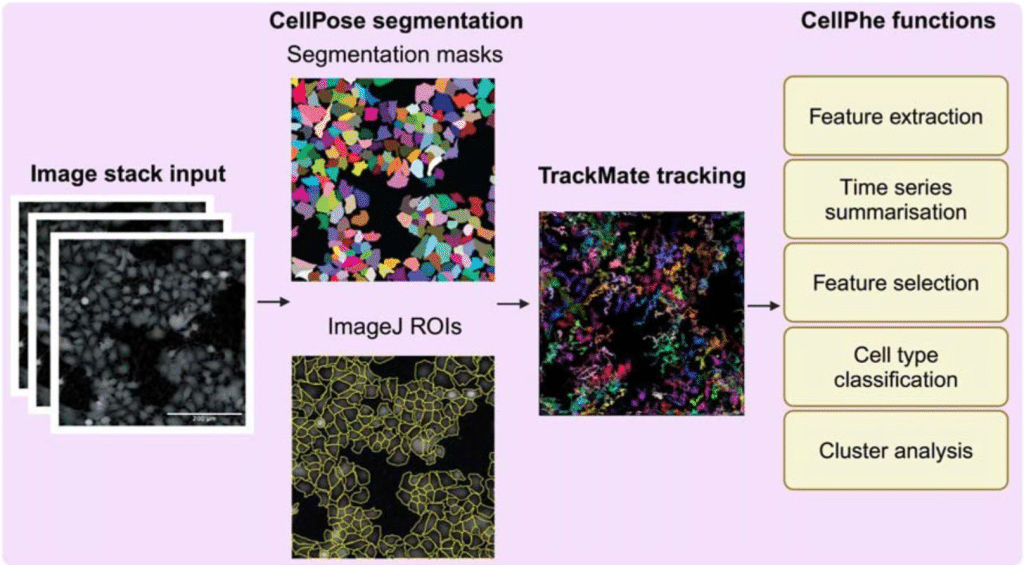
Microscopy Nodes: versatile 3D microscopy visualization with Blender
Oane Gros, Chandni Bhickta, Granita Lokaj, Yannick Schwab, Simone Köhler, Niccolò Banterle
HD2Net: A Deep Learning Framework for Simultaneous Denoising and Deaberration in Fluorescence Microscopy
Xuekai Hou, Yue Li, Chad M. Hobson, Hari Shroff, Min Guo, Huafeng Liu
Systematic analysis of immune cell motility leveraging Immunemap, an open intravital microscopy atlas
Diego Ulisse Pizzagalli, Pau Carrillo-Barbera, Elisa Palladino, Kevin Ceni, Benedikt Thelen, Alain Pulfer, Enrico Moscatello, Raffaella Fiamma Cabini, Johannes Textor, Inge M. N. Wortel, The Immunemap project consortium, Rolf Krause, Santiago Fernandez Gonzalez
Active Prompt Tuning Enables Gpt-40 To Do Efficient Classification Of Microscopy Images
Abhiram Kandiyana, Peter R. Mouton, Yaroslav Kolinko, Lawrence O. Hall, Dmitry Goldgof
Zero-Shot Image Denoising for High-Resolution Electron Microscopy
Xuanyu Tian, Zhuoya Dong, Xiyue Lin, Yue Gao, Hongjiang Wei, Yanhang Ma, Jingyi Yu, Yuyao Zhang
A Novel Variational Approach for Multiphoton Microscopy Image Restoration: from PSF Estimation to 3D Deconvolution
Julien Ajdenbaum, Emilie Chouzenoux, Claire Lefort, Ségolène Martin, Jean-Christophe Pesquet
CryoVIA – An image analysis toolkit for the quantification of membrane structures from cryo-EM micrographs
Philipp Schönnenbeck, Benedikt Junglas, Carsten Sachse
Ilastik: a machine learning image analysis platform to interrogate stem cell fate decisions across multiple vertebrate species
Alma Zuniga Munoz, Kartik Soni, Angela Li, Vedant Lakkundi, Arundati Iyer, Ari Adler, Kathryn Kirkendall, Frank Petrigliano, Bérénice A. Benayoun, Thomas P. Lozito, Albert E. Almada
DeepPaint: A deep-learning package for Cell Painting Image Classification
Diego Luna, Erik C. Johnson, Laura J. Dunphy, Ryan McQuillen
FastReseg: using transcript locations to refine image-based cell segmentation results in spatial transcriptomics
Lidan Wu, Joseph M. Beechem, Patrick Danaher
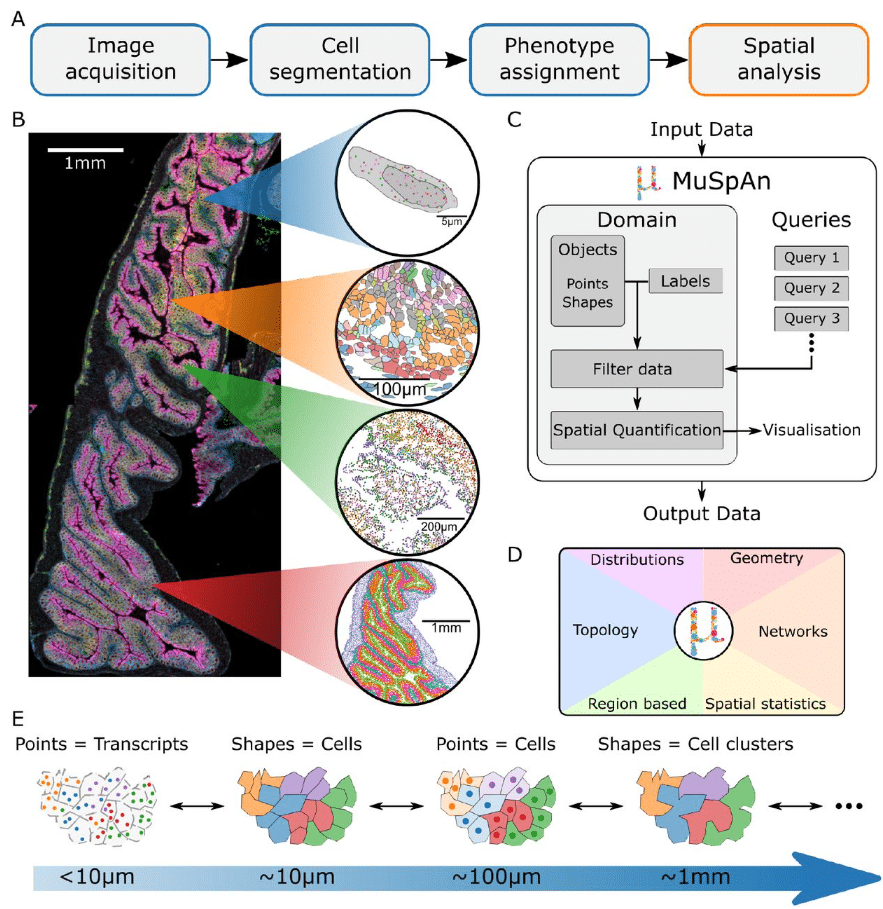
MuSpAn: A Toolbox for Multiscale Spatial Analysis
Joshua A. Bull, Joshua W. Moore, Eoghan J. Mulholland, Simon J. Leedham, Helen M. Byrne
MerQuaCo: a computational tool for quality control in image-based spatial transcriptomics
Naomi Martin, Paul Olsen, Jacob Quon, Jazmin Campos, Nasmil Valera Cuevas, Josh Nagra, Marshall VanNess, Zoe Maltzer, Emily C Gelfand, Alana Oyama, Amanda Gary, Yimin Wang, Angela Alaya, Augustin Ruiz, Cade Reynoldson, Cameron Bielstein, Christina Alice Pom, Cindy Huang, Cliff Slaughterbeck, Elizabeth Liang, Jason Alexander, Jeanelle Ariza, Jocelin Malone, Jose Melchor, Kaity Colbert, Krissy Brouner, Lyudmila Shulga, Melissa Reding, Patrick Latimer, Raymond Sanchez, Stuard Barta, Tom Egdorf, Zachary Madigan, Chelsea M Pagan, Jennie L Close, Brian Long, Michael Kunst, Ed S Lein, Hongkui Zeng, Delissa McMillen, Jack Waters
Impact of Segmentation Errors in Analysis of Spatial Transcriptomics Data
Jonathan Mitchel, Teng Gao, Eli Cole, Viktor Petukhov, Peter V. Kharchenko
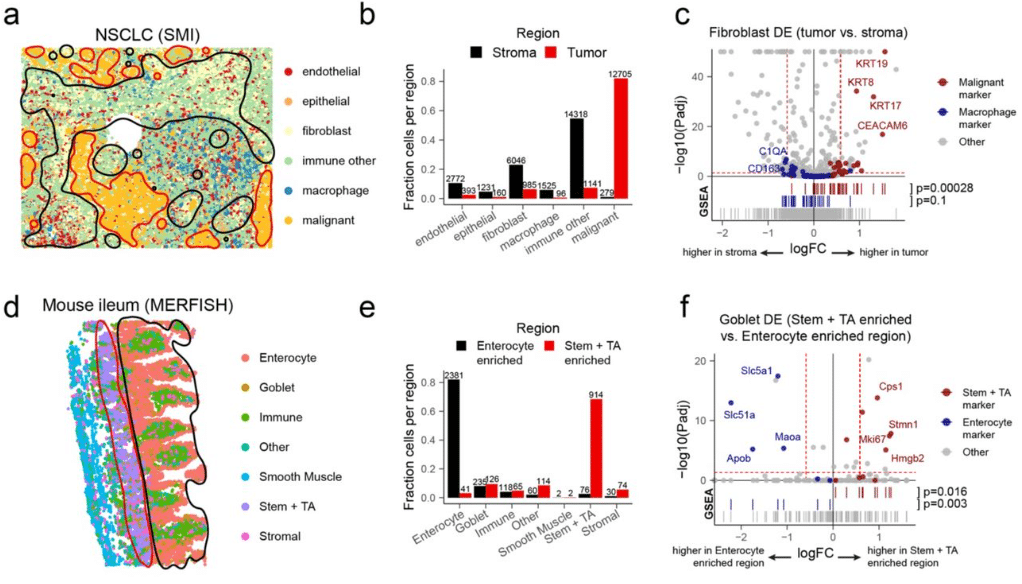
Assessing large multimodal models for one-shot learning and interpretability in biomedical image classification
Wenpin Hou, Qi Liu, Huifang Ma, Yilong Qu, Zhicheng Ji
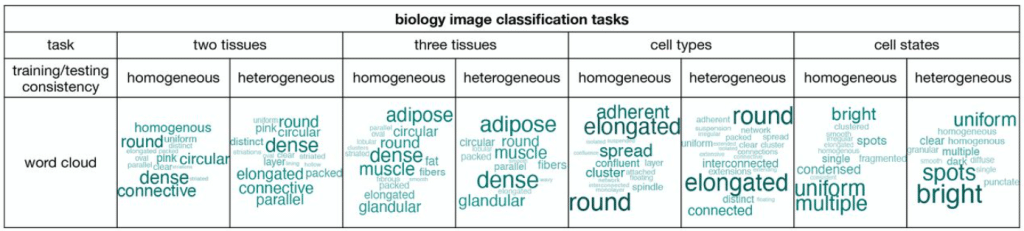
Accessible and accurate cytometry analysis using fluorescence microscopes
Daniel Foyt, Yiming Kuang, Samma Rehem, Klaus Yserentant, Bo Huang
A Study on Context Length and Efficient Transformers for Biomedical Image Analysis
Sarah M. Hooper, Hui Xue
SEW: Self-calibration Enhanced Whole Slide Pathology Image Analysis
Haoming Luo, Xiaotian Yu, Shengxuming Zhang, Jiabin Xia, Yang Jian, Yuning Sun, Liang Xue, Mingli Song, Jing Zhang, Xiuming Zhang, Zunlei Feng
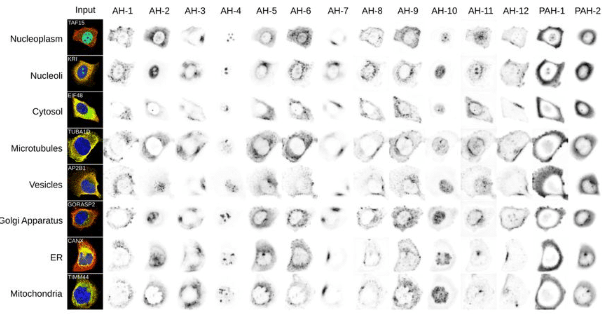
SubCell: Vision foundation models for microscopy capture single-cell biology
Ankit Gupta, Zoe Wefers, Konstantin Kahnert, Jan N. Hansen, Will Leineweber, Anthony Cesnik, Dan Lu, Ulrika Axelsson, Frederic Ballllosera Navarro, Theofanis Karaletsos, Emma Lundberg


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)