TrackMate-Oneat: Auto Track correction using deep learning networks
Posted by varun kapoor, on 4 July 2022
During the tracking of motile cells, solving the problem of linking objects between two consecutive timepoints becomes even more complicated, if the cells divide or undergo cell death. In the terms of trajectories, this means the addition of trajectory branches and terminations. However, dividing and dying cells are characteristic in their shape, and leveraging this biological information can aid the detection of cell division and death events, making cell tracking much more accurate. In this article we present our TrackMate action called TrackMate-Oneat (made by Varun Kapoor (Kapoorlabs, Paris), Mari Tolonen (University of Copenhagen, Denmark), a tool to autocorrect lineage trees by leveraging prior information about the cell events detected via a neural network such as oneat. The cell events supported in this action include mitosis and cell death, knowing the location of these events we autocorrect cell lineages using a local Jaqaman linker and also leverage the geometrical prior for angular location of daughter cells with respect to the mother cell. We detail the usage of this action along with other tools required to create the files needed for this action.
Installation instructions
For installation of the python library please see the instructions on it’s github page for installation: Installation Instructions. For installation of the TrackMate extension please click on the update site of TrackMate-Oneat to get the updated jar in your Fiji installation.
A detailed explanation of the algorithms behind TrackMate-Oneat can be found here TrackMate-Oneat wiki on Fiji.
Prerequisites for usage
As a prerequisite for using the tool you will need to have an integer labelled segmentation image for your dataset as the track correction requires precise information about the cell location. You can use our segmentation plugin in Napari called VollSeg-Napari to obtain a VollSeg1 segmentation of your cells in 2D/3D using pre-trained or your own StarDist/CARE/U-Net models: Segmentation models.
Subsequently, you need to have a trained oneat model for your dataset. We have several pre-trained models available that you can use on similar datasets. You can download the available pre-trained models from here: Pre-trained models. Once you have an oneat model you can apply it on a 3D + time/2D + time dataset of arbitrary size image using this notebook: Jupyter notebook code.
Solving the non maximal suppression in time
A curse of the neural networks is over-detection of events the network is trained to find, especially when the network is fully convolutional. This happens because each pixel in the image has an opinion about the event classification with a certain probability. Such over-detection is however nuisance, as it leads to detection of event slightly before or after it actually happens. To avoid this, we created a Napari widget that inputs the .csv file of the cell event locations, along with the segmentation image and allows the users to interactively veto the events above a certain threshold, which we apply in a certain time block (also a user adjustable parameter) to obtain a non maximally suppressed detection of only the mother cells, since in our model training the mother cell was in the central frame and central XY location, the probability score at the mother cell frame tends to be higher than at frames before and after. Using this notebook Jupyter Notebook with Napari widget we accomplish the task of performing non maximal suppression in time.
This video Walkthrough the Napari widget is a walkthrough in using this Napari widget.
Tracking in TrackMate
Finally, the information about cell division and death locations are combined with particle linking using TrackMate2 in ImageJ3. The linking of particles themselves is achieved using the TrackMate LAP (Linear Assignment Problem) Tracker. The users have to create a hyperstack in ImageJ with the Raw image in one channel and the integer labelled image in the other channel and then use the integer label detector of TrackMate for the spot detection process (Fig. 1). After the detection process the users can choose the Simple LAP tracker that ´does not create any prior cell division links and only outputs cells linked to each other over frames. Otherwise the users can choose the LAP tracker that creates links between the segments and we can use Oneat correction with both modes of tracking (Fig. 2).
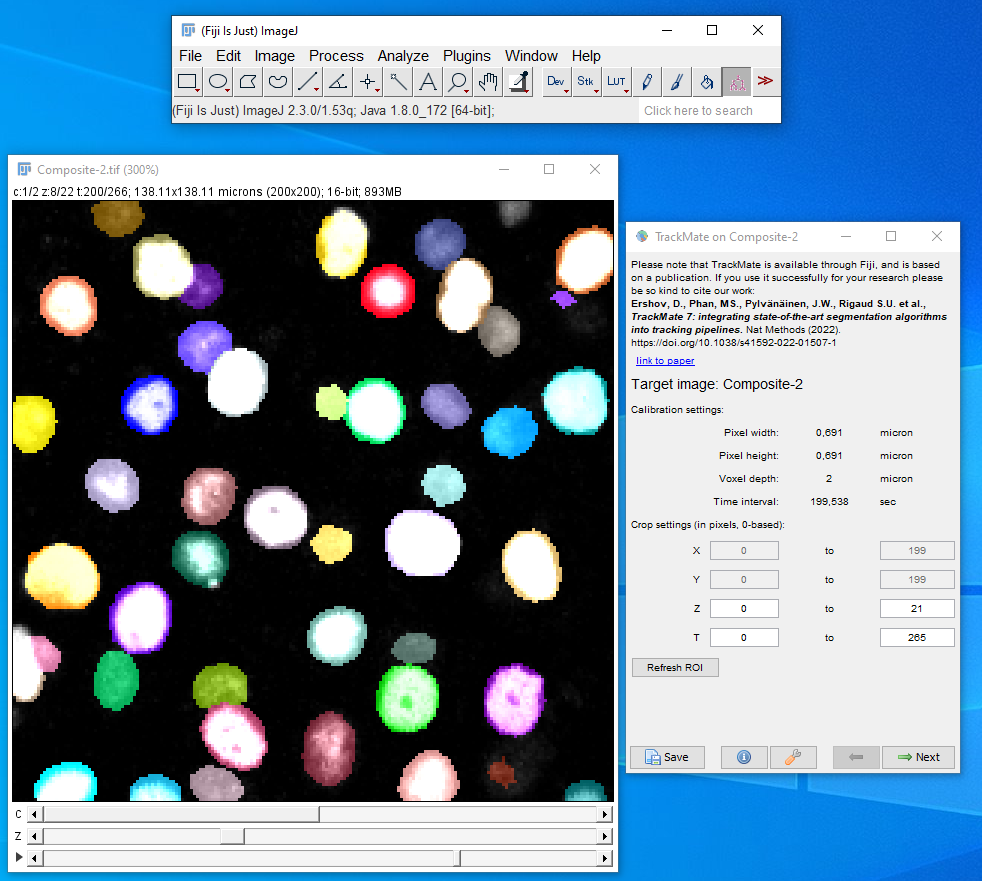
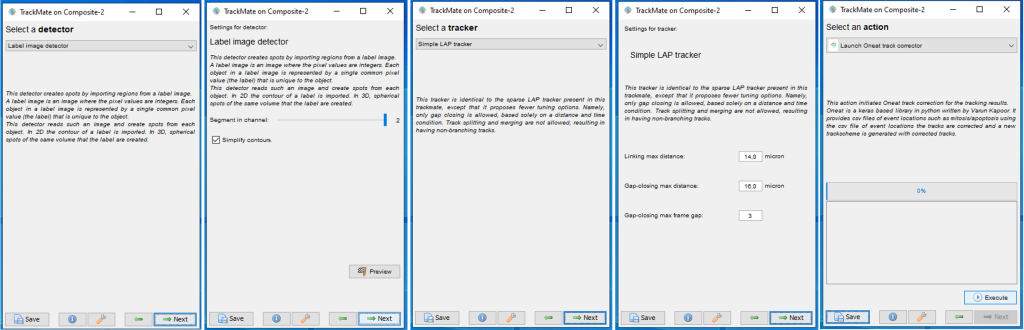
After the users reach the last panel, select the TrackMate-Oneat action from the panel that opens the TrackMate-Oneat corrector (Fig. 3). In this panel, the users can load the clean .csv file of cell division and death detections (This file we obtained after performing the non maximal suppression in time in the previous step in Napari). Furthermore there is a time parameter which considers a TrackMate found link to be correct if its location lies in the temporal neighborhood of it, else the link would be broken by the Oneat corrector. The distance threshold in space we use to look for the tracks to link to the mother cell locations found by oneat is the same as used in the Simple LAP/LAP step of the tracking process.
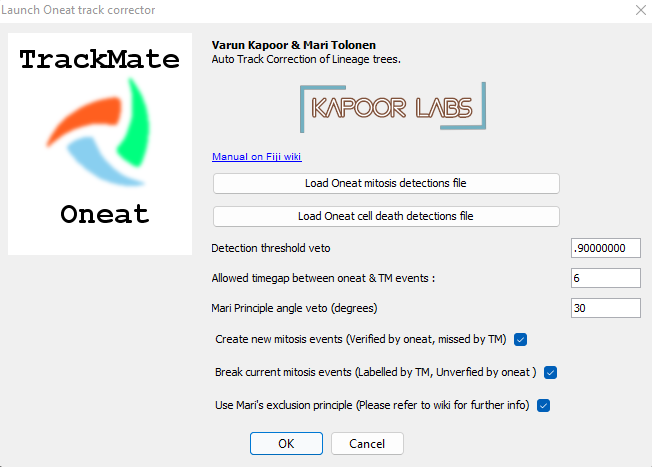
Using cell fate information in track linking
The output of oneat is a set of locations in time of the mother cells, hence for cell tracks we can use this information to find the mother cells, and to create links between the mother and the daughter cells in the next frame. For this, we search for the candidate tracks in the next frame in a defined region and solve local Jaqaman cost matrix4 to create links between the mother cell track and the likely assignments. Such an assignment still can go wrong due to erroneous links arising after minimizing the cost matrix. Fortunately we can further minimize the error of creating such links using biological and physical prior as described in the next section.
Using principles of biology and physics in track linking
Besides having the prior information of cell events in terms of the location and frame of the mother cells, we also have built in the biological prior and prior based on physics to auto correct the lineage trees. This is based on the geometrics of labelled nuclei during mitosis. For all mitotic events, the mother cell acquires a characteristic shape during metaphase, where the chromatids are assembled on a metaphase plate, usually in the middle of the cell. As the cell divides, the daughter chromatids are pulled by the mitotic machinery to the opposite sides of the cell.5 When the cell divisions are symmetric, i.e. the cell division occurs on the same cell layer, the daughter chromatids can thus be seen moving in exactly opposite directions. Thus, cell division proceeds along the minor axis of an ellipse fitted around the contour of the segmented nucleus (Fig. 4).
When the cell division events are added into solving the cost matrix, the physical principles of momentum conservation can be applied to this phenomenon. Knowing that the cell division proceeds with only two and not as three or more daughter cells implies that the angles, theta and phi of the daughters wrt the minor axis of the ellipse are small (Fig. 5). By invoking the principle of momentum conservation on top of the biological prior enforces a further constraint in finding the daughter cells, which is on the opposite side of the major axis of the ellipse as is illustrated by the momentum balance equation in Fig. 5. This further constrains the search zone of finding the daughter cell to be on the opposite side of the major axis. This video explains these principles in more detail: Tutorial on Youtube
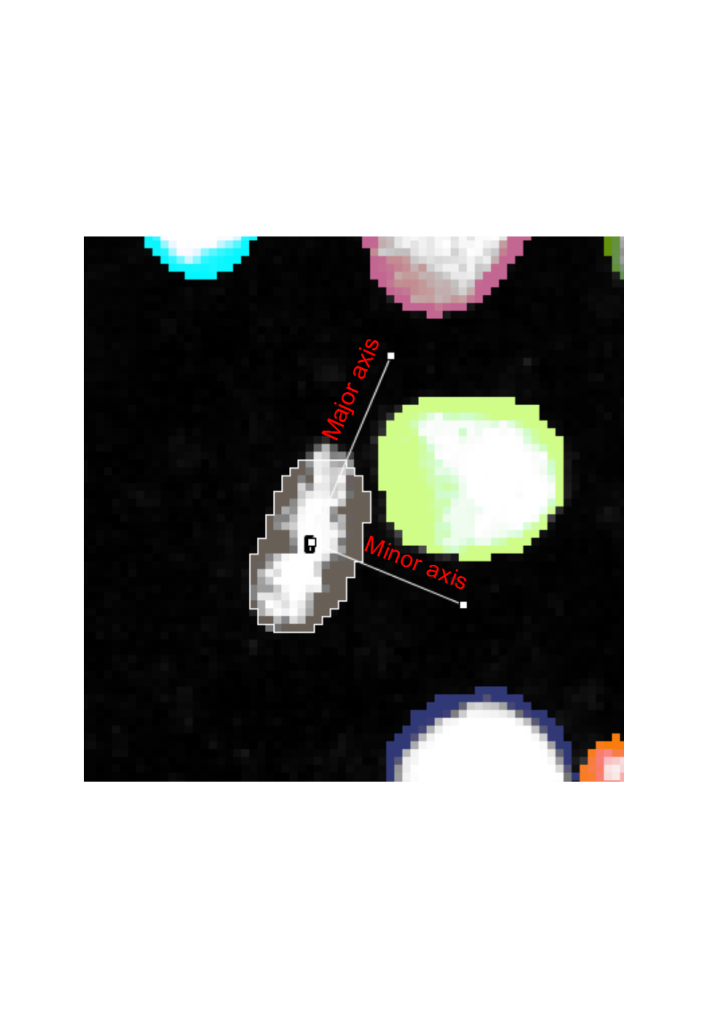
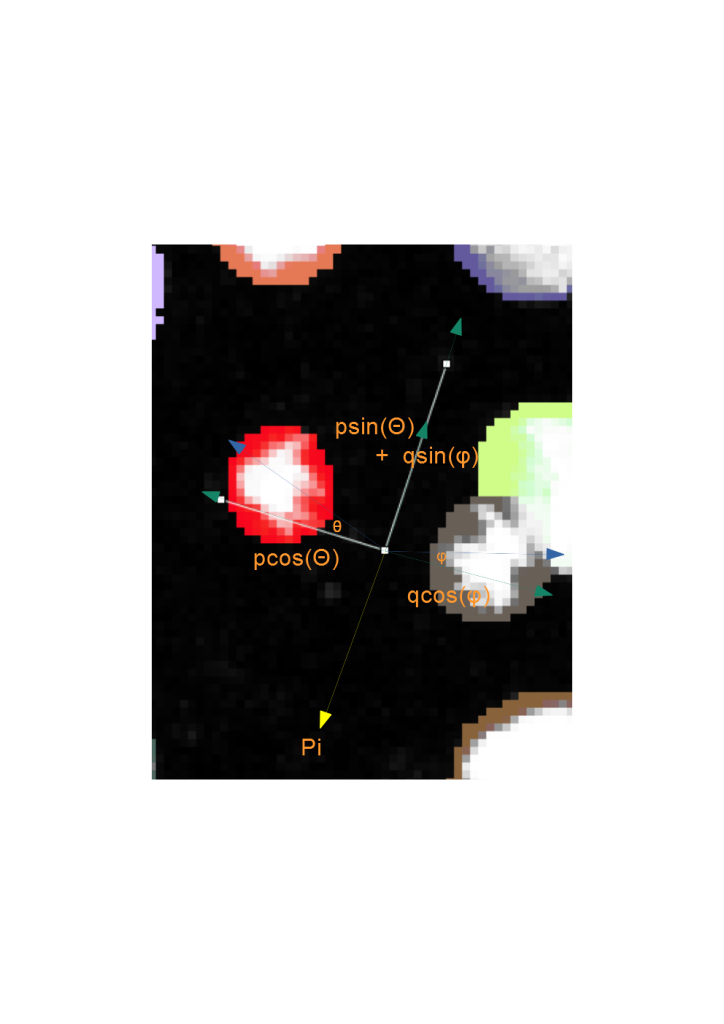
For the biological organisms that do not necessarily adhere to the biological motion observed, (e.g., if cell divisions happen asymmetrically, and thus daughter cells don’t move to the opposite directions in the field of view), the user can increase the search angle to 90 degree, in which case the only the daughter cells that lie on the opposite sides of the major axis would be linked. For particles that do not adhere to these principles (e.g. blebs or other non-mitotic events), this principle can be disabled. In the TrackMate-Oneat action panel we call this principle as Mari’s principle, as it was inspired by Mari Tolonen and programmed by Varun Kapoor in imglib2 for this action.
Support
- The image.sc forum is the best place to start getting help and support. Make sure to use the tag
oneat, since we are monitoring all questions with this tag. - If you have technical questions or found a bug, feel free to visit our Github page to open an issue.
References
- Kapoor, V., & Carabaña, C. (2021). Cell Tracking in 3D using deep learning segmentations. Python in Science Conference, (pp. 154-161). Austin (virtual), United States.
- Ershov, D., Phan, M.-S., Pylvänäinen, J. W., Rigaud, S. U., Le Blanc, L., Charles-Orszag, A., . . . Tinevez, J.-Y. (2022). TrackMate 7: integration state-of-othe-art segmentation algorithms into tracking pipelines. Nature Methods.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., . . . Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676-682.
- Jaqaman, K., Loerke, D., Mettlen, M., Kuwata, H., Grinstein, S., Schmid, S., & Danuser, G. (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods 5 (8), 695-702.
- Gadde, S., & Heald, R. (2004). Mechanisms and Molecules of the Mitotic Spindle. Current Biology 14 (18), R797-R805.


 (6 votes, average: 1.00 out of 1)
(6 votes, average: 1.00 out of 1)
