Enhancing Global Access: interview with CZI grantee Uri Manor
Posted by Mariana De Niz, on 24 January 2024

OPEN DISSEMINATION OF NOVEL IMAGING TOOLS FOR THE RESEARCH COMMUNITY
Uri Manor is an imaging scientist with a background in cell biology and computational methods. His project focuses on building new probes to improve and accelerate research, and on using artificial intelligence to break barriers in the way we do microscopy.
What was the inspiration for your project? How did your idea for the CZI project arise?
I highlighted work that was in progress at the time: the generation of probes, and AI-based image analysis. When the call came out, I felt it was tailored for someone like me – It was supporting all the type of work I was doing, as a Core Director and as a scientist. Sometimes people have to try very hard to fit into the mold of funding agencies, but in this case it was a natural fit. Hopefully CZI feels the same way!
In terms of AI and the probes you designed, how did you realize this was something necessary for the community?
To me it’s obvious that you need probes to be able to visualize anything at a microscope. There are some exceptions as is the case with label-free microscopy and cryo-EM, but in most other cases, we struggle with probes that are not as specific as we would like them to be, as well as other challenges. At the microscope, you can only see what you put on, so sample preparation itself is a major part of the art of microscopy, and probe design is part of this art. In terms of AI, we are constantly making decisions and tradeoffs. We sometimes struggle at the microscope because in the imaging universe you have to choose between speed or resolution or signal and sample preservation. That’s something that every imaging scientist knows intrinsically. Having free software that can run in any computer with a descent GPU, to make possible what is otherwise impossible – to me this is an obvious and exciting direction to pursue.

The idea of integrating AI to the microscope itself is great. Can you tell us more about it?
Technologies have always been an opportunity for us to improve our quality of life, and to do things we couldn’t do before. I see self-driving microscopes as an example. For instance, when we are trying to capture rare events (either in time or space) – it would be great to have a self-driving microscope that can monitor the landscape and then recognize when a rare event is happening (or about to happen), and then zoom in and capture the event. This would get us much higher throughput! And I think it’s really necessary because there is so much variability in biology. Being able to increase the end values for so called rare events is super important. Another thing we are working on is correlative microscopy. The highest resolution we can achieve with imaging is via cryo-EM or EM. We can get atomic or near-atomic resolution of biological structures, but it’s so laborious and it takes so long to get just one dataset. Moreover, that dataset is a frozen snapshot of one time point. However, one of the great things about biology is that it’s dynamic and it changes, so it’s almost impossible to describe a biological system using only a static one-timepoint picture of it, without showing how dynamic it is. For example, mitochondrial fission is something I’ve been studying for a long time. We would love to have an atomic resolution picture of what happens during this process. We can do that with cryo-EM to a certain extent, but we really need to build a movie – we need to capture fission events at different time points and in different contexts. A self-driving AI microscope could learn how to know when a mitochondrial fission event is occurring, and where it’s occurring. It could be attached to a high-pressure freezer or a microfluidic device that could fix the sample at different stages of fission, and tell us where the event is, so that we can take the sample, put it in the cryo-EM and build up that dynamic picture using static approaches. With the CLEM idea, one thing we are working on, is correlating data to dynamic movies. There are organelles moving around the cell and when we fix the sample, we know exactly what was moving where, at the time of fixation. We can then do EM in those samples, and annotate those same images with the movement that we captured in the video, and then train an AI to be able to predict the movement from the EM image. We are working on all this. It’s very low throughput at the moment, but I think this is revolutionary: having a microscope that can automatically communicate with a freezing device so that we can then do EM, and once we have EM, we annotate it for more AI to do other things. We can use AI to take microscopy to a whole new level. However, one of the challenges that come with smart microscopes, is making them accessible. One of my big goals – which I hope to accomplish by the end of my CZI tenure, is to make user-friendly interfaces so that people can use AI algorithms without needing to install complicated combinations of libraries and code.
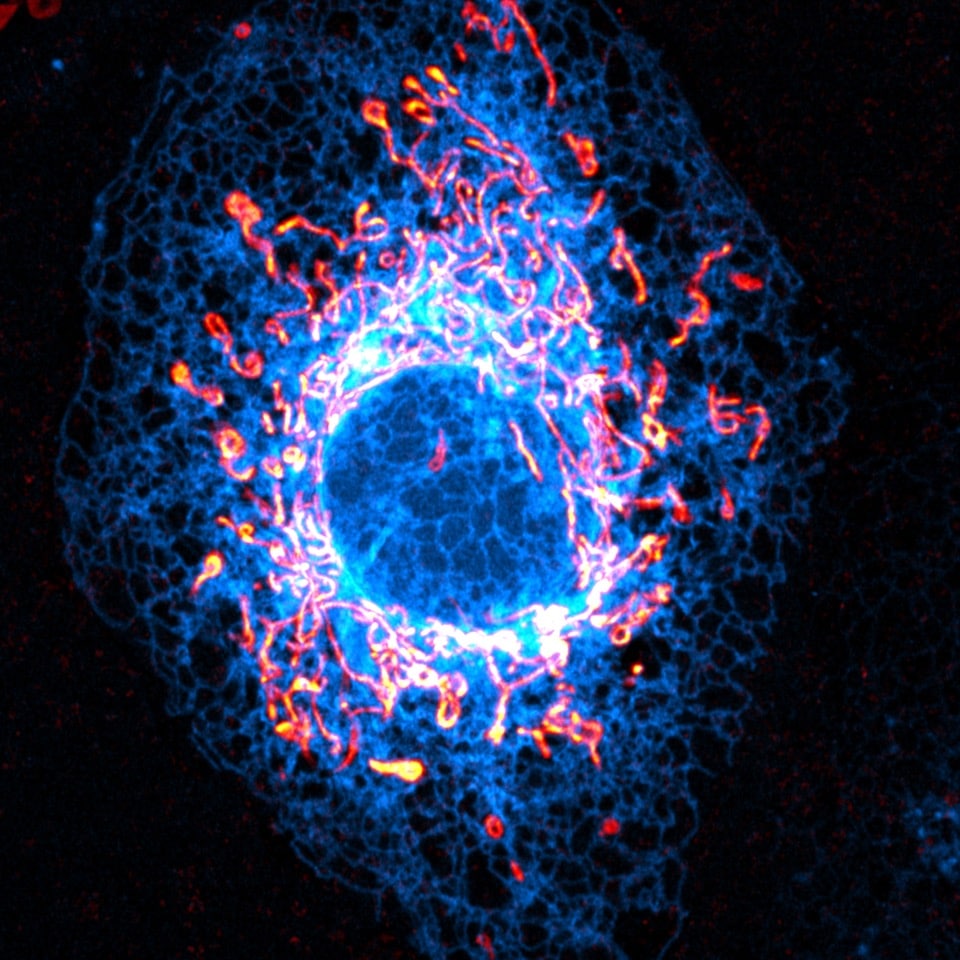
You are a big supporter of open access, open science and open dissemination. What do you think is the importance of openness?
My dream world is that everything is open – our microscopes, our lab meetings, etc: anyone can just come along and look at what we are doing, and immediately start making decisions and using the knowledge that is being generated for their own research, in real time. Take Twitter for example: people figured out that one doesn’t need to wait for a story to be written in a newspaper. Sometimes you could find out about world events right away. I think there are analogues between journalism and science: you witness and then you report, and I would love to see science happening at the speed of Twitter (or maybe a better platform). This to me is utopia, and sadly we don’t live in utopia. On one hand, your collaborators might not feel the same way about open science or preprints. It’s almost paradoxical though, that a true open scientist collaborates with everyone, but part of collaborating means respecting the threshold of privacy and other values or opinions of your colleagues, which might conflict with open science. This is something that I’m still learning how to navigate. Even in my own lab, I don’t want to be a dictator that says ‘we only publish preprints’. I think everyone has a perspective of value and I want to be able to simultaneously maintain my principles and honour others’. Going back to my idea of utopia and fast communication speed: a problem with immediate science is the same as Twitter: Twitter moves at the speed of light but so does the misinformation that travels in it. So in science, if you show every result of every experiment in real time and then realize something was an artefact, you could contribute to wasted time and efforts. There’s a reason why we value rigour and reproducibility, and why we don’t publish everything straight away. It’s a balancing act! But in terms of publishing, I think eLife got it right with the post-publication peer review. That’s actually how the human race has always functioned: we share stories, we experience life, and then we figure out which stories were correct and which ones weren’t. And this is constantly being updated. Theories in Physics and Biology are updated all the time as we encounter mountains of data to refute our old models and we have to rebuild them. I feel the publication process is artificial and in many ways distorted – it’s a source of tension, distorted science, and gatekeeping. Actually, Sam Lord, from Dyche Mullins lab recently put out a blog on what the gold standard for publications should be. I’ve been thinking about this too, for a long time: a journal system where you send your results for them to be reproduced. The journal’s staff job is to reproduce whatever you report. Because otherwise, not because something is written does it mean it’s true, even with peer-review. And peer review itself is not without its own dangers. True anonymity is basically impossible because it’s easy to pinpoint where certain work comes from. For reviewers, sadly, lack of anonymity is still linked to possible repercussions, especially for young scientists. I think everything would be better if science was preprinted!
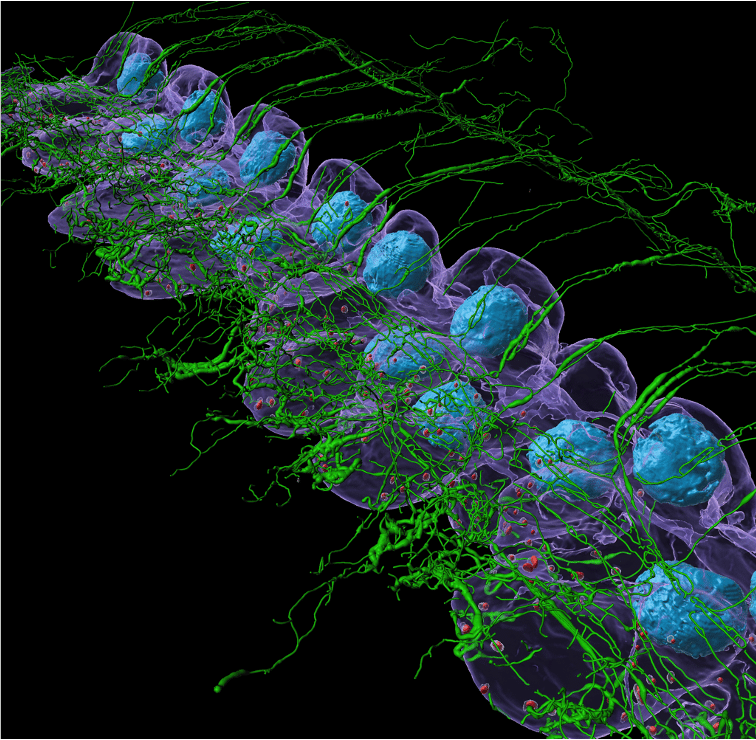
How instrumental was the CZI grant itself in allowing you to execute your ideas?
Infinitely instrumental. I’m not sure it would have been doable. I was able to do things before CZI, but not at the level I can now. Funding is everything -it’s the difference between being able to hire someone to help and get work done, so you can work on other things and together move forward faster and better. There is also the issue of gatekeeping: when I got the award, I feel my peers saw me a little bit differently, and the institute saw me a little bit differently too. They were more willing to support me and to accommodate me, which has been a longstanding problem for core facility scientists and technology developers in general. Historically, people focused in technology have been given a lot less respect by their biology peers. This never made sense to me, I think there is huge value in technology development. So for me the CZI award was a turning point.
What have been the biggest successes and the biggest challenges of your project?
The biggest success was being able to put more time and energy into AI research. Also, I’ve shared the nanobody probes with over 20+ labs and this is a great example of win-win open science. Actually it was during the preprint stage of my work that I started getting requests for the constructs and I started shipping them out. Several collaborations which have culminated in publications, have been possible since, and this, in my opinion, is a big success. In terms of the AI work, we published an AI segmentation paper recently on local shape descriptors. We have more work on the pipeline, addressing bottlenecks for AI. Not everyone has access to it, or can generate it, so we are working on an approach that will allow you to generate segmentation with much less training data. In terms of challenges, it’s difficult and expensive to generate software that is biologist-friendly. I want some really slick software that is appealing and easy to use, but so far I haven’t achieved this.
You do a lot in terms of teaching. How do you feel teaching can bridge gaps between disciplines and maybe make AI more accessible altogether?
I am now at UCSD, and part of my job now in addition to directing, is running educational workshops. I’ll be teaching courses to undergraduate students. I have support from UCSD to build a brand new course that has never been taught before, which is image analysis for undergraduate students. What I intend to do is teach cell biology from a microscopy and image analysis point of view. I’m going to go through the cell biology curriculum, and for every single organelle or signaling pathway or process that is taught in cell biology, I’m going to find a microscopy experiment that proves or disproves the model and I’m going to give them the students this data. They are then going to analyse the images from those microscopes to help them draw biological conclusions from the data. So they’ll learn different things: how imaging reveals processes, what is the suitable control, etc. I’m anxious because I have never taught this course. I also want to produce something to get it up on YouTube and make it open and accessible to everyone. But I don’t know if UCSD has rules about making courses for free. I have a lot to learn, but I’m excited about it.
How or why did you choose to apply to CZI and what have you learned about the CZI program overall, that you previously didn’t know? And what would you change if you could?
It was very easy to apply. There were no other programs for supporting core staff and/or technology developers the way this call has. This was the only grant I ever heard of, which I was eligible for. This is otherwise a big problem in science- that so many awards are only for tenured track professors or equivalent. By funding our work, CZI showed that our work is valuable. I think CZI’s philosophy is my soulmate in terms of funding – it’s all about open science and doing big things and making maximal impact. They just announced a new round of Ben Barres early investigator grants, they have a CZI diversity program – they are doing everything right, in my opinion. CZI seems to also really value open source, and they incentivize it. And of course, I’m biased, but I love that they chose to support imaging scientists! We need it! There are some things I would change about the imaging scientist grant: I think it should be given to the individual, not the institute. Also, think about how much more impactful an imaging scientist grant could be if there was also budget for equipment. The most creative imaging scientist work will usually involve some kind of new instrument that can be developed. Finally the other thing I have brought up in the past is software engineering. Often, a software engineering problem is best solved by a seasoned software engineer who can work on a project for focused time, as opposed to hiring and training someone new to figure out how to do things – this takes longer and is not done as successfully. Developing software funding would be great, so that we can hire experts or outsource projects, or to have a team of software engineers who can work with CZI imaging scientists to help bring software to production so it’s stable, usable, friendy, dowloadable and compatible with any operating system. This would be cool.
How do you feel CZI has fostered community building?
When I’m at a CZI meeting, I do feel I’m with my people, almost more than in any other meeting. It’s really nice, and they all care, and they are excited about the same thing, so those meetings are very special. That said, meetings are tough: it’s hard to travel, hard to get around, and they are very intense. You can only handle so much interaction. The issue with meetings is that we should find a way to reinforce and maintain the connections that we make during them. Everyone would appreciate an efficient way to do this.
How important is to you the idea of democratizing microscopy?
The people I admire the most are people who have produced a probe they placed on Addgene, which has been downloaded 500,000 times. The point of tools is to use them, and the point of our effort is to make a difference in the world. Democratizing means that the most amount of people will get to use a tool we build. I want my tools to reach as many people as possible and to have as much impact as possible. However, while chatting with you, I wonder about the word ‘democratize’ itself: democracy means everyone has an equal voice, but what we really want is broad distribution. Accessibility is a better word for me. I want everybody to have equal access to tools.
Final remarks: I hope CZI sees our gratitude and realizes the enormity of the impact of what they have done, and that they choose to invest more in it, and that they are proud of what they have done.
Check out our introductory post, with links to the other interviews here


 (No Ratings Yet)
(No Ratings Yet)