Featured image with Ludovica Altieri
Posted by FocalPlane, on 15 August 2025
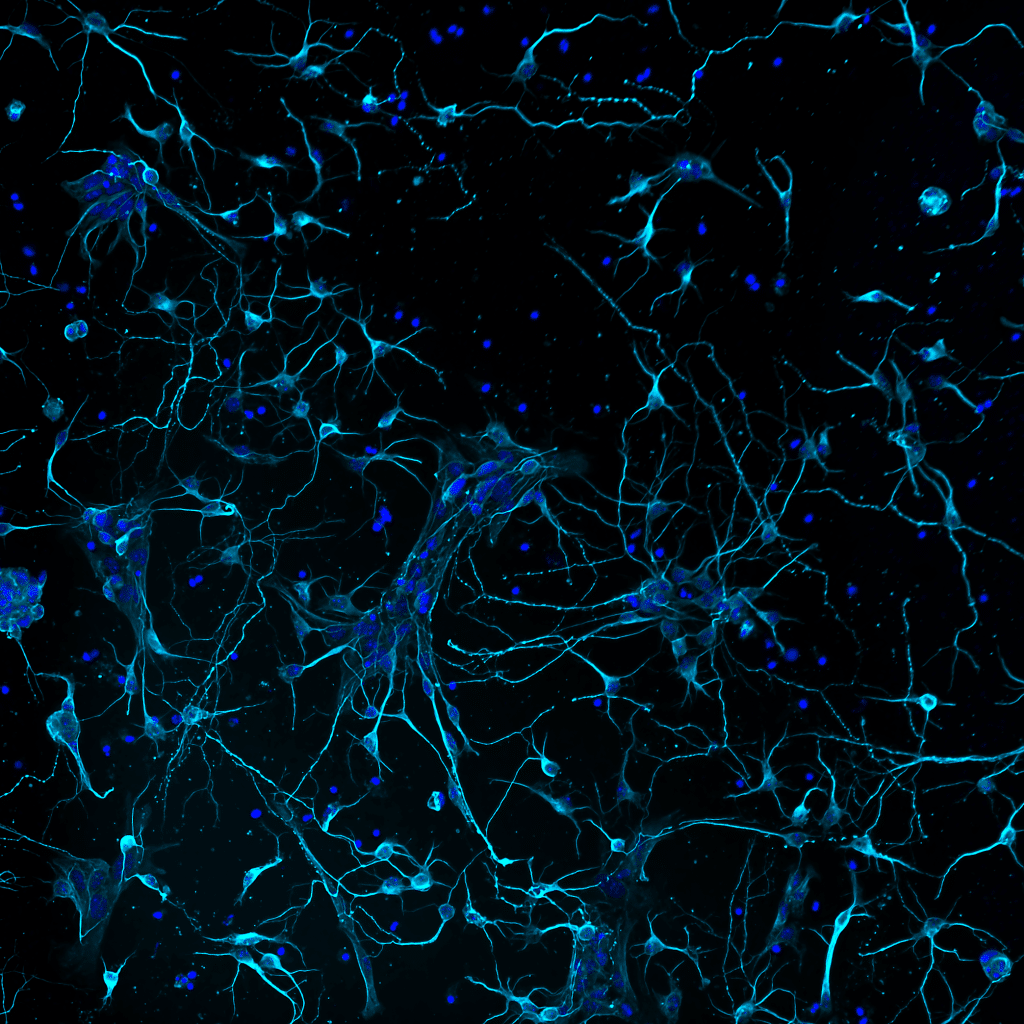
Our featured image, acquired by Ludovica Altieri, shows murine primary cortical neurons developing interconnections. The cells are stained for neuronal tubulin (TUJI antibody, cyan) and DNA (DAPI, blue). Cultures were prepared by dissecting cortices from mouse embryos (E18), then mechanically dissociating single cells for plating. Explanted primary neurons placed in culture regress several stages back, then, when cultured in appropriate medium, they spontaneously resume differentiation over time during Days in vitro (DIVs). The imaged sample was taken at DIV 4 using a Nikon microscope implemented with a CrestOptics confocal spinning disk module with post-processing using NIS Elements AR by Nikon.
Discover more about Ludovica’s research:
Research career so far: I have recently obtained my PhD degree in Genetics and Molecular Biology at Sapienza University of Rome, Italy. In my studies I explored the roles of mitotic regulators in microtubule dynamics, both during mitosis and in neurodifferentiation. I am a cellular biologist by training, with imaging expertise spanning from quantitative immunofluorescence of fixed samples to time-lapse video recording of living cells and high-content imaging pipelines. I am fascinated by the potential of these approaches in studies of cell dynamics, cell architecture and biological networks.
Current research: I am currently an early career post-doc researcher at the Institute of Molecular Biology and Pathology – CNR National Research Council of Italy with Professor Patrizia Lavia in the framework of the Italian national microscopy infrastructure to develop innovative imaging methods. Most processes in biology are dynamic, yet most commonly used techniques miss out on that dimension. My current focus is the role of mitotic genes in neurodevelopmental disorders such as microcephaly, a failure of proper of brain growth. I am using imaging, a combination of quantitative microscopy, in vivo time-lapse video recording and biochemical techniques, to pinpoint the role of specific genes in neurodifferentiation, e.g. neurite maturation, primary cilium formation, axon pathfinding and dynamics. I am also characterising tubulin modifications, and the enzymes involved, in cancer cells, as well as studying bacterial strains entry into, and infection of, mammalian cells via plasma membrane remodeling.
Favourite imaging technique/microscope: I find DeepSIM super resolution microscopy, which I have experienced at CrestOptics, really powerful. DeepSIM is designed to work with samples of comparable thicknesses to those used in confocal microscopy, giving super-resolved data over 50 μm-depth in nonclarified samples, such that heterogeneous, complex samples can be thoroughly investigated. Also, as a visitor at the Italian Institute of Technology, I have become acquainted with 3D cell systems, brain organoid technologies and 3D bioprinting. These techniques, applied to 3D cell systems, have an amazing power in studies of CNS models.
What are you most excited about in microscopy?
In addition to the techniques mentioned above to image dynamic processes, upcoming challenges will require dissecting biological processes at nanoscale to resolve the subcellular structures and molecules in the buildup of organelles and whole cells. To that aim, ExM, Cryo-EM and 4D-STEM will prove instrumental to achieve full understanding of complex cellular processes.


 (No Ratings Yet)
(No Ratings Yet)