Creating a Research Data Management Plan using chatGPT
Posted by Robert Haase, on 6 November 2023
TL;DR: Data Management Plans (DMPs) are documents which describe what happens to data in a [research] project. More and more funding agencies require these documents when scientists apply for funding. However, different funding agencies may require different information in DMPs. In this blog post I will demonstrate how chatGPT can be used to combine a fictive project description with a DMP specification to produce a project-specific DMP. You can reproduce my steps using the Jupyter Notebook published in this repository.
What is a DMP about?
Before we dive into writing down details, we ask chatGPT 4.0 for a basic DMP document structure. It will likely list similar sections like shown in this screenshot:
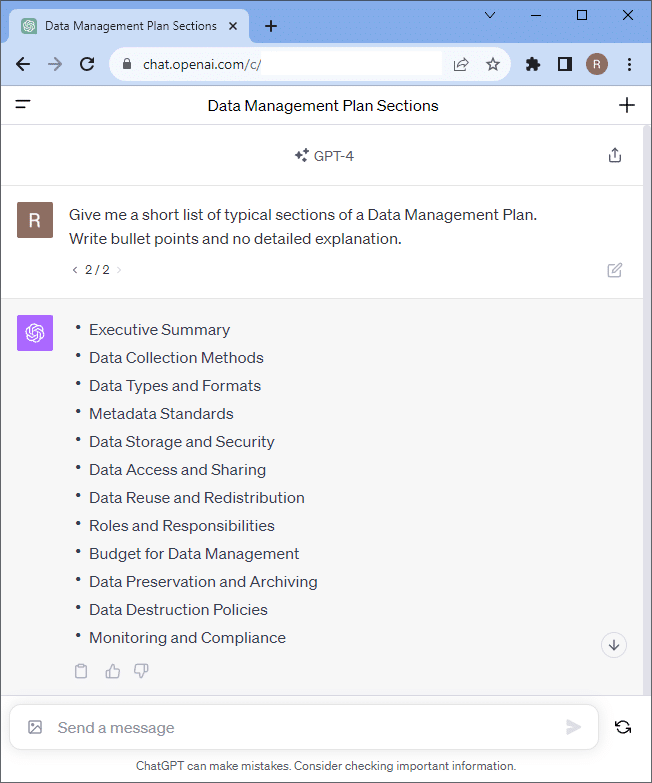
You can also ask for details and explanations of specific sections.
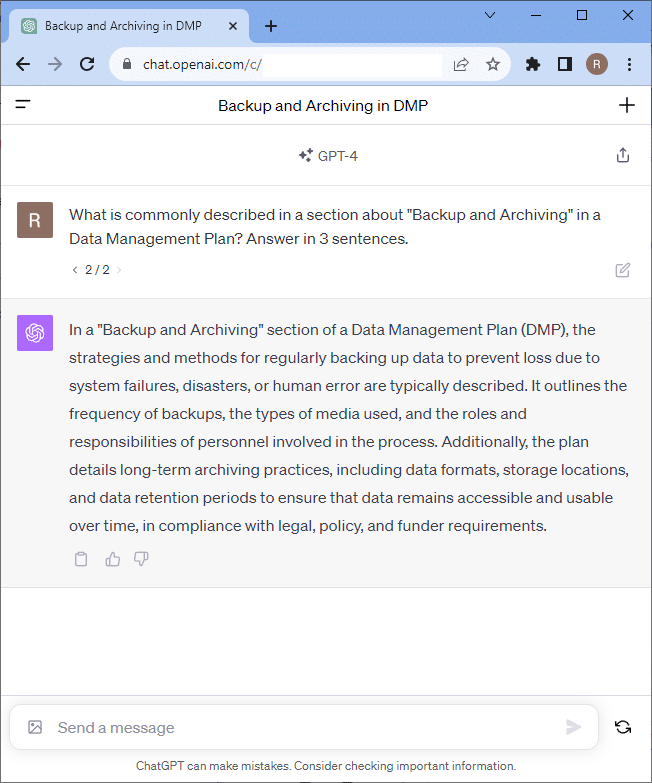
Our project
With this overview of sections and some knowledge of your specific project, you can write a text describing your typical workflow in projects. You could then ask chatGPT to turn your project description into a DMP. When writing your project description, keep in mind that you must comply with what you describe in your DMP. The document is not the right place to describe how your ideal project looks like. I recommend describing the way how you deal with data reflecting reality. If you for example describe using some infrastructure you do not have access to, you will later have a hard time arguing that your project went according to the plan. This is my unstructured description of a fictive project:
In our project we investigate the underlying physical principles for Gastrulation in Tribolium castaneum embryo development. Therefore, we use light-sheet microscopes to acquire 3D timelapse imaging data. We store this data in the NGFF file format. After acquistion, two scientists, typically a PhD student and a post-doc or group leader look into the data together and decide if the dataset will be analyzed in detail. In case yes, we upload the data to an Omero-Server, a research data management solution specifically developed for microscopy imaging data. Data on this server is automatically backed-up by the compute center of our university. We then login to the Jupyter Lab server of the institute where we analyze the data. Analysis results are also stored in the Omero-Server next to the imaging data results belong to. The Python analysis code we write is stored in the institutional git-server. Also this server is backed up by the compute center. When the project advances, we start writing a manuscipt using overleaf, an online service for collaborative manuscipt editing based on latex files. After every writing session, we save back the changed manuscript to the institutional git server. As soon as the manuscript is finished and submitted to the bioRxiv, a preprint server in the life-sciences, we also publish the project-related code by marking the project on the git-server as public. We also tag the code with a release version. At the same time we publish the imaging data by submitting a copy of the dataset from the Omero-Server to zenodo.org, a community-driven repository for research data funded by the European Union. Another copy of the data, the code and the manuscript is stored on the institutional archive server. This server, maintained by the compute center, garantees to archive data for 15 years. Documents and data we published is licensed under CC-BY 4.0 license. The code we publish is licensed BSD3. The entire project and all steps of the data life-cycle are documented in an institutional labnotebook where every user has to pay 10 Euro per month. Four people will work on the project. The compute center estimates the costs for storage and maintenance of the infrastructure to 20k Euro and half a position of an IT specialist. The project duration is four years.
As you can see, this text contains plenty of information about the research data life-cycle of our project. But various aspects are mixed. I wrote about file formats, responsible people and IT infrastructure in a mixed way, pretty much how it came to my mind while describing the project in a chronological way. We will now use chatGPT to turn this wildly structured project description into a DMP.\
A first DMP
In the following, I will use chatGPT via the OpenAI API to enable readers to more easily reproduce my steps and potentially adapt them to their needs. You can download the notebook I used to prompt chatGPT and adapt it to your needs. In this notebook you will also see the questions I asked above and that the answer chatGPT is different from the screenshots above. This is quite common when using large language models. The technology produces results influenced by randomness.
A prompt to chatGPT commonly starts with a scenario description instructing the language model what kind of task comes next. Note that {project_description} is replaced by my project description above.
You are a professional grant proposal writer. In the following comes a description of
a common project in our "Tribolium Development" Research Group at the University.
Your task is to reformulate this project description into a Data Management Plan.
{project_description}And this is the response:
Data Management Plan for Investigation of Gastrulation in Tribolium Castaneum Embryo Development
- Data Collection: Data is procured through the use of light-sheet microscopes, which produces 3D timelapse imaging data, archived in the NGFF file format. The data review is performed by a minimum of two researchers, following which a decision on detailed data analysis is made.
- Data Storage: If the data is chosen for further analysis, it is uploaded to an Omero-Server, a platform specifically designed for microscope imaging data management. The platform is backed up by the university’s compute center, ensuring the data’s security. Analysis results are stored alongside the original data for easy access and reference.
- Data Processing: Data processing is carried out on the institute’s Jupyter Lab server. Python is used to code the analysis, which is then stored on the institutional git-server, offering another level of data backup.
- Documentation: The team documents every stage of the project in an institutional lab notebook. The manuscript drafting is conducted via Overleaf, an online collaborative tool, with every version being saved back into the institutional git-server.
- Data Availability: When complete, manuscripts are submitted to bioRxiv, a preprint server in life sciences. The code relative to the project is made public on the git-server and tagged with a release version. Imaging data is submitted to zenodo.org, a EU-funded research data repository. Additionally, a complete copy of project materials is stored on the institutional archive server for a guaranteed period of 15 years.
- Licensing: All published documents and data fall under the CC-BY 4.0 license, while the published code is licensed BSD3.
- Personnel: A team of four is assigned to the project. The university’s compute center also dedicates half an IT specialist’s role to this project for maintenance and support.
- Costs: Estimated costs for storage and infrastructure maintenance are roughly 20,000 Euros, with an expected project duration of four years.
- Compliance: All team members are required to adhere to the guidelines set within this data management plan.
Combining information and structure
If a funding agency expects a DMP in a specific format, you should respect this format and tell chatGPT about it. In the following example, I will use guidelines from the German Research Foundation (DFG) published in 2021. DFG’s Checklist Regarding the Handling of Research Data contains these sections, which we can re-use to formulate our DMP:
- Data description
- Documentation and data quality
- Storage and technical archiving the project
- Legal obligations and conditions
- Data exchange and long-term data accessibility
- Responsibilities and resources
Thus, you can make use of chatGPT to reformulate your project description into a specific format, that might be different from project to project. We modify the prompt from above and add information about the structure we need for our DMP, and we specify that the text should be formatted using markdown enabling structured headlines.
You are a professional grant proposal writer. In the following comes a description of
a common project in our "Tribolium Development" Research Group at the University.
Your task is to reformulate this project description into a Data Management Plan.
{project_description}
The required structure for the data management plan, we need to write is like this:
# Data Management Plan
## Data description
## Documentation and data quality
## Storage and technical archiving the project
## Legal obligations and conditions
## Data exchange and long-term data accessibility
## Responsibilities and resources
Use Markdown for headlines and text style.And that was its output:
Data Management Plan
Data Description
Our study aims at understanding the fundamental physical principles informing Gastrulation in Tribolium castaneum embryo development. Through this endeavor, we will generate 3D timelapse imaging data gathered from light-sheet microscopes. The data sets are stored using the NGFF file format. In the process of the project, we will create Python-based code for data analysis, research papers drafted on Overleaf, which incorporates the findings and results of our study.
Documentation and Data Quality
Once the required data is acquired, it undergoes a scrutiny process where two scientists, generally a post-doc or group leader along with a PhD scholar, determine whether the data merits a detailed analysis. If affirmed, it will be stored on our Omero-Server – an exclusive data management solution created for microscopy imaging data. The analysis results, Python analysis scripts, and manuscript edits are also stored in this server and always updated after each modification.
Storage and Technical Archiving of the Project
The data on the Omero-Server and the institutional git-server, where we store the Python analysis code and edits to our manuscript, are automatically backed up by our university’s compute center. Completed aspects of the project are retained in the institutional archive server for a guaranteed period of 15 years. This archive likewise holds a copy of all published data, code, and manuscripts attached to the project.
Legal Obligations and Conditions
Our published documents and data are licensed under the CC-BY 4.0 license, while the published code follows the BSD3 license. Individuals working on the project and accessing the institutional lab notebook are required to pay a monthly fee of 10 Euros.
Data Exchange and Long-term Data Accessibility
To ensure wider accessibility and visibility, we publish our finalized manuscripts to the bioRxiv – a preprint server primarily geared towards the life-sciences. We further open-source our project-related Python code by switching the settings on our git-server to public and tagging the code with a release version. All imaging data will be made accessible by uploading a copy of the data set from the Omero-Server to zenodo.org – a community-driven repository for research data backed by the European Union.
Responsibilities and Resources
The research group comprises four members who will be actively involved in the project that spans over four years. The compute center estimates the infrastructural costs for both storage and maintenance to be 20k Euros and one-half of an IT specialist’s position.
Limitations
Tools based on large language models such as chatGPT work obviously quite well for re-organizing information and reformulating text. However, if you do not provide information for specific sections, it may come up with random proposals for these sections. It may also loose information you provided. You obviously need to read the document before submitting it to the funding agency. By the way, DFG wrote in a statement in 2023 that using chatGPT for writing proposals is fine and the submitting scientist remains responsible for the content of the document. Thus, again, you need to read carefully the DMP you submit, and later you will have to follow the plan during the project. You may want to mention that chatGPT was used for bringing your DMP into the right format though.
Further reading and who to talk to
Many universities and research institutes provide more information about DMPs on their websites. Some insitutions have founded service units where so called Data Stewards work who can support you when writing DMPs. For example, a good starting point to search for information in your institution might be contacting the colleagues who work in the library. Many libraries have undergone a transition from a place where books are stored to a place where people meet to learn more about how to deal with data in the digital age. In Germany, you find National Research Data Initiatives (NFDIs) such as NFDI4BioImage, where I am happy member of, who can provide field specific support in handling research data. Reach out, all NFDIs are open-minded groups of people who are happy to help, e.g. when writing DMPs.
In the project description above I mentioned licenses and repositories for publishing code and data. You can read more about those in these blog posts:
- Sharing research data with Zenodo
- Sharing Your Poster on Figshare: A Community Guide to How-To and Why
- If you license it, it’ll be harder to steal it. Why we should license our work
- Collaborative bio-image analysis script editing with git
Conclusions
Large language models like chatGPT transform the way we do science, including how we apply for projects and report about them. If you use the technology to write a DMP, this is certainly no guarantee that your project gets funded. But it may help formatting the DMP the right way. I hope this short guide is useful and I’m happy about feedback.
Reusing this material
This blog post is open-access, figures and text can be rused under the terms of the CC BY 4.0 license unless mentioned otherwise.


 (3 votes, average: 1.00 out of 1)
(3 votes, average: 1.00 out of 1)