See more with HCR spectral imaging
Posted by FocalPlane, on 6 March 2024
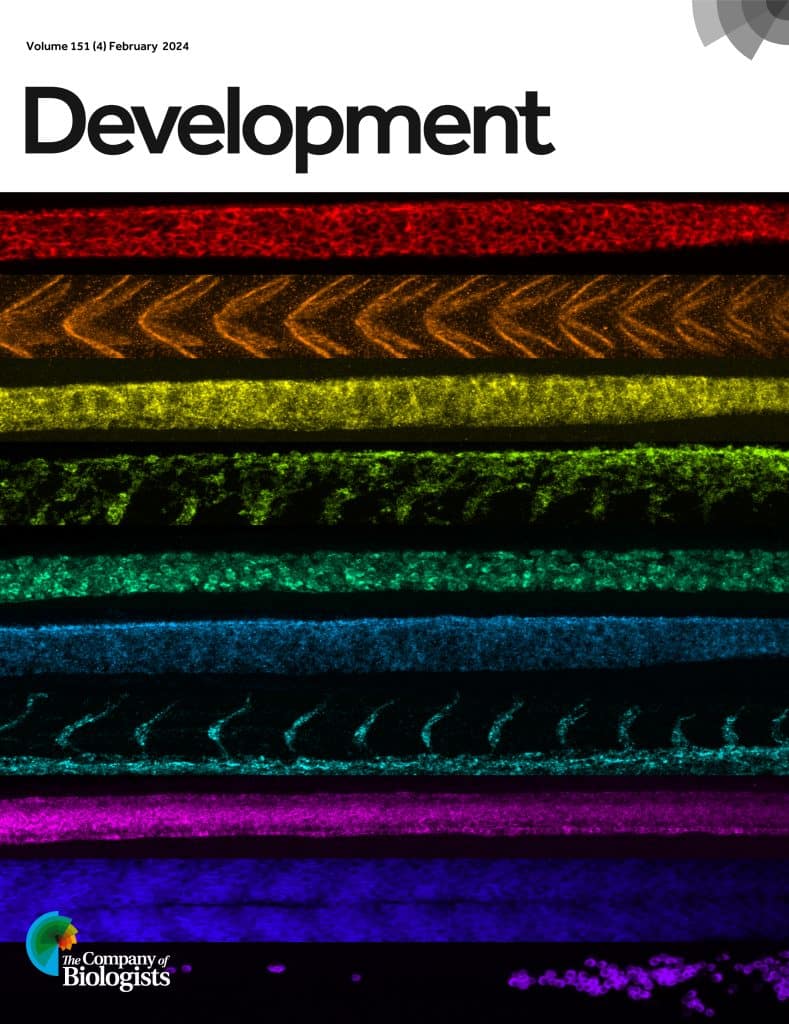
Optimizing labelling is essential for improving signal to noise ratio when imaging thick samples with high autofluorescence. One method of achieving this is using hybridization chain reaction (HCR) to amplify the signal in a quantitative manner. HCR can be combined with RNA fluorescence in situ hybridization (FISH) to label RNA or with immunofluorescence to label proteins. In a new paper published in Development, Samuel Schulte, Niles Pierce and colleagues present a new workflow for HCR spectral imaging with linear unmixing, for simultaneous imaging of ten RNA and/or protein targets in both whole-mount zebrafish embryos and mouse brain sections. They design an optimized set of ten orthogonal HCR amplifiers to be used in combination with ten fluorophores with spectra that can be separated by linear unmixing. The linear unmixing algorithm requires reference spectra for each fluorophore plus reference spectra for the autofluorescence. The robust protocol should allow researchers with the necessary imaging hardware to replicate the quantitative imaging in samples with high autofluorescence.
To read more about HCR from the Pierce lab, check out the following papers in Development:


 (No Ratings Yet)
(No Ratings Yet)