Imaging spotlight: Lock-in-SIM
Posted by FocalPlane, on 8 January 2026
In this paper highlight from Wenjie Liu and colleagues, we learn about Lock-in-SIM, a new methodology for structured illumination microscopy.
Can you briefly describe Lock-in-SIM?
Lock-in-SIM is an open-access two-dimensional structured illumination microscopy (SIM) reconstruction framework. Inspired by the classical lock-in amplifying detection approach used in electronic circuits to separate the background noise from the signal, Lock-in-SIM exploits the modulation differences of the periodic SIM illumination pattern for sample structures of varying whole-cell depths to lock, extract, and amplify the weak high-frequency signal deriving from subdiffraction-sized sample features while minimizing the effects of unmodulated background. By eliminating background, Lock-in-SIM achieves the generally opposing goals of high spatio-temporal resolution, high signal-to-background-ratio, 3D optical sectioning, whole-cell-depth, artifact-free, and high-fidelity imaging.
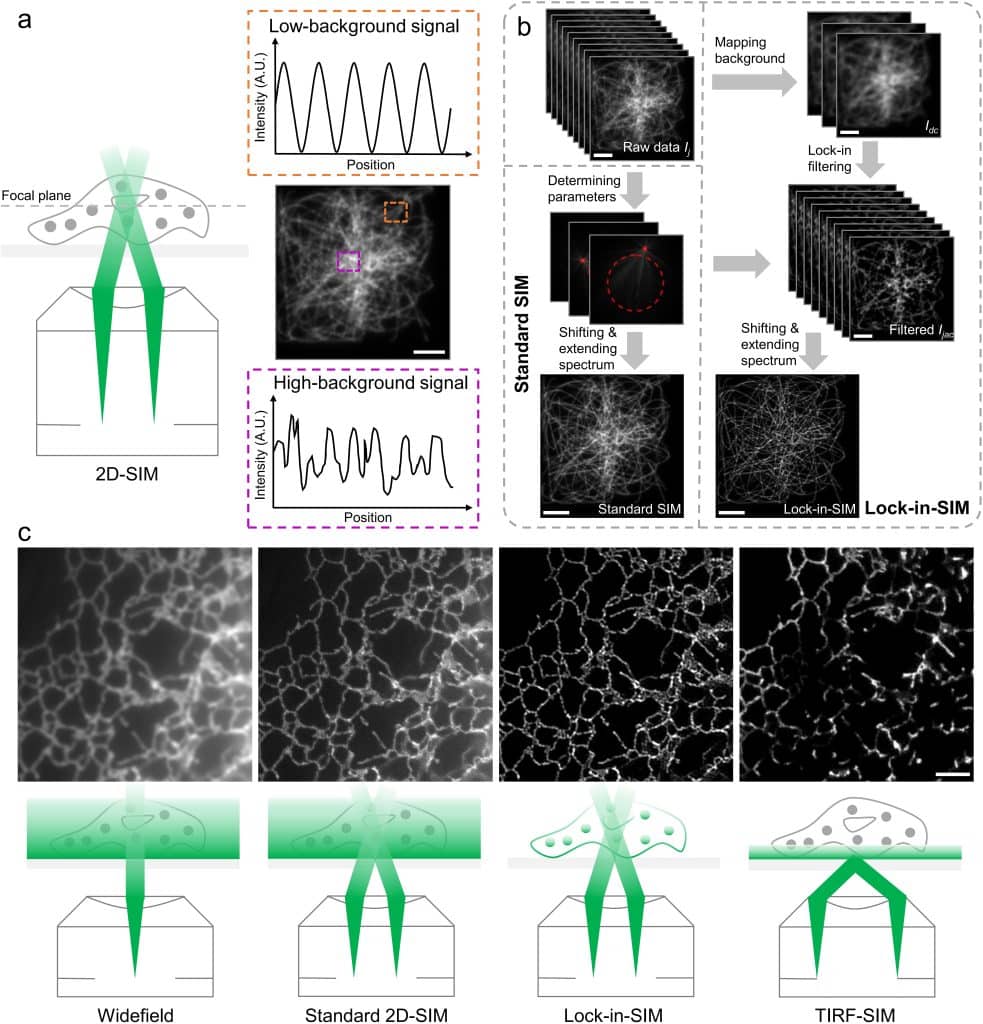
How has Lock-in-SIM been used for so far?
The generalized implementation of Lock-in-SIM enables the background- and artifact-free visualization of a wide range of fixed and live subcellular and intraorganellar ultrastructures with the highest resolution, fidelity, and imaging depth theoretically attainable with SIM. Through quantitative long-term live-cell imaging, we demonstrate an improvement in quantification sensitivity and a reduction in analysis bias by utilizing Lock-in-SIM reconstruction. In two-color time-lapse ER-lysosome imaging, we successfully captured the structural rearrangements of intraorganellar components, including the ER matrix and lysosome tubules, and their interactions. In multi-color time-lapse mitochondria-dynamin-related protein 1 (Drp1) imaging, we reveal a mechanism of mitochondria-mediated mitochondrial fission via the transfer of Drp1, completing the classical mitochondrial fission model based on other subcellular components and shedding light on the interactions of organelles. Lock-in-SIM combines compelling reconstruction quality with universal applicability, in a user-friendly open-access package, that will greatly facilitate the exploration of highly intricate and dynamic subcellular processes.
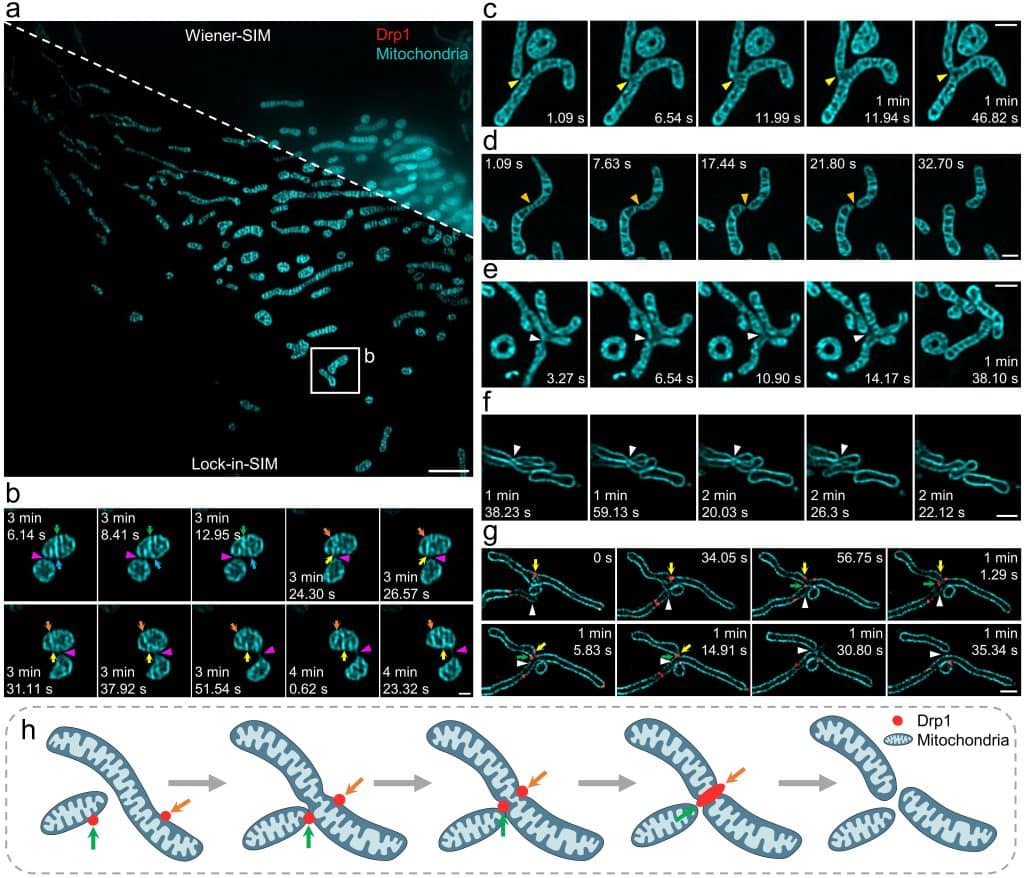
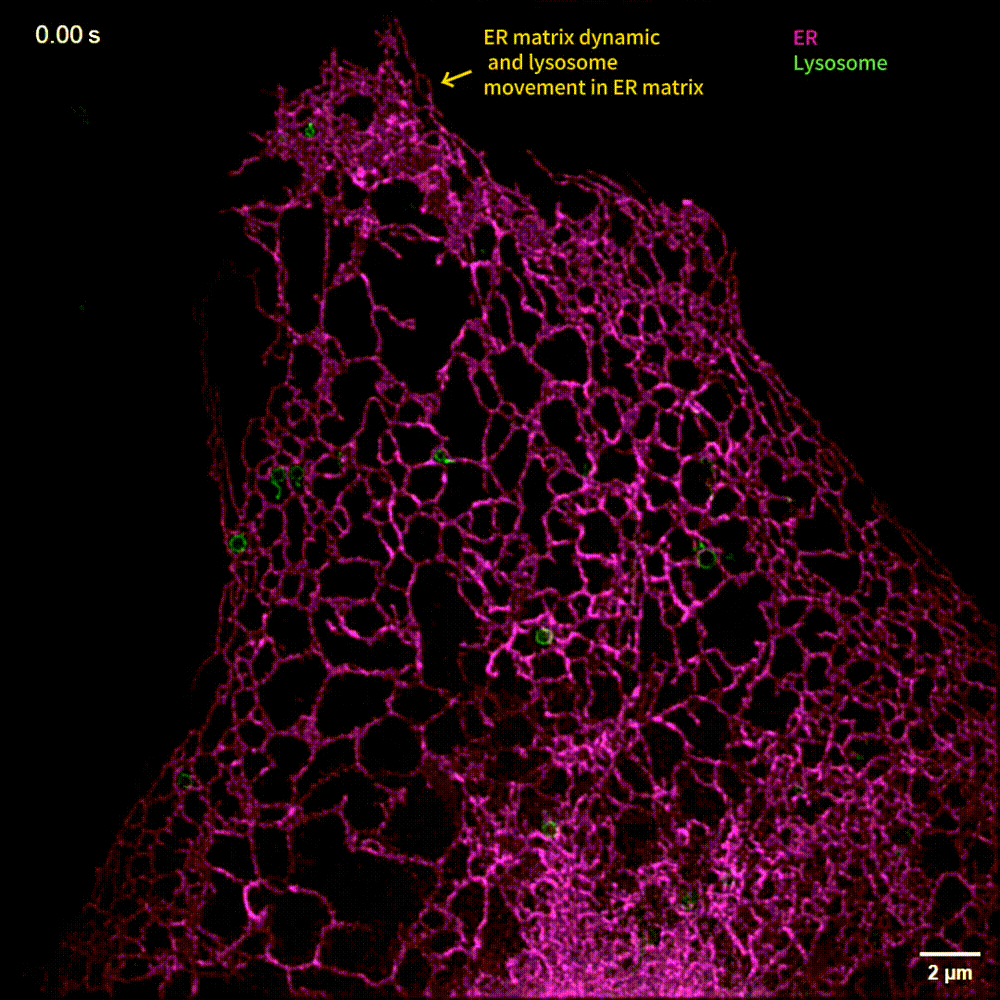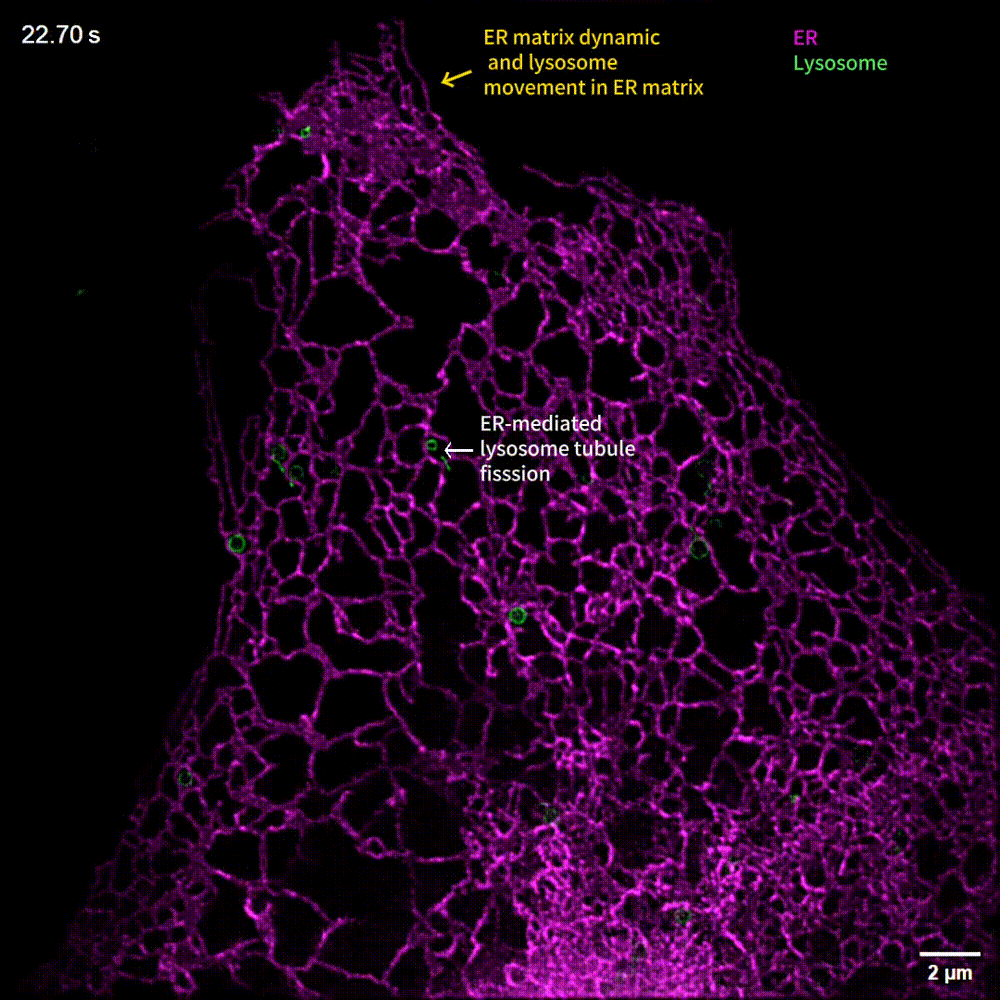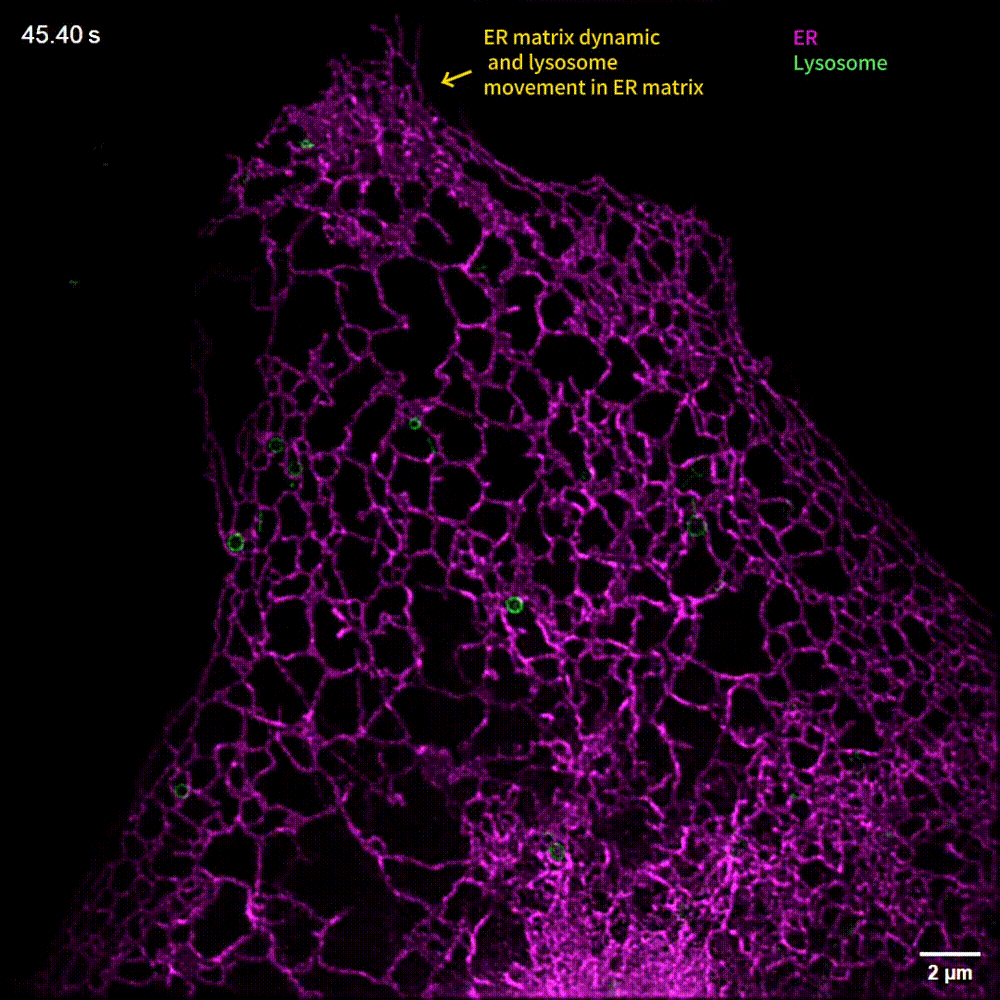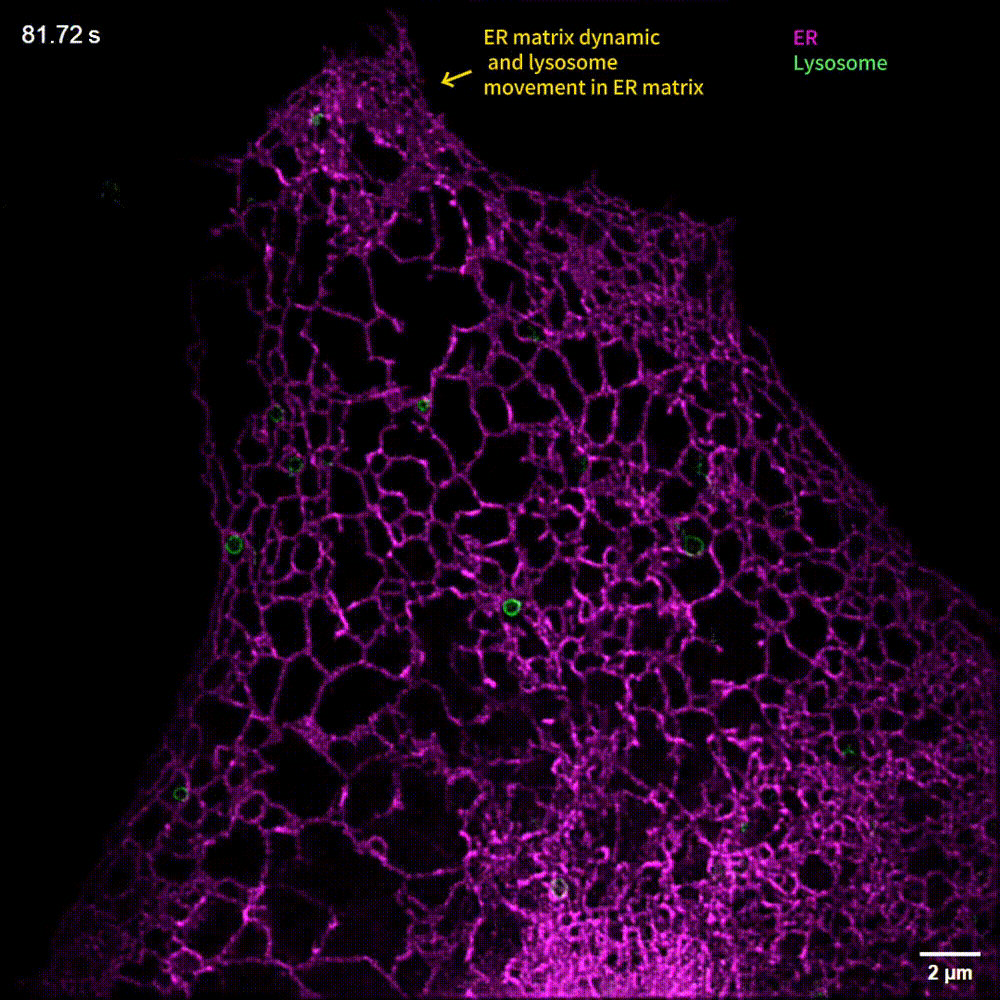
What do researchers require to implement Lock-in-SIM?
We offer three open-access Lock-in-SIM software platforms: MATLAB GUI, Fiji/ImageJ plugin, and an executable software version for widespread use of our method. This is the first open-access algorithm compatible across these three platforms and the second Fiji/ImageJ plugin in 2D-SIM field. As Lock-in-SIM process is inherent in standard SIM imaging and can therefore be applied to data acquired from existing SIM systems directly, allowing for seamless integration into established post-processing workflows without additional hardware modifications and troublesome parameter fine-tuning. To facilitate users of commercial SIM systems, we additionally provide a Fiji/ImageJ plugin for converting various SIM data sequences, enabling also seamless integration of our algorithm into diverse commercial 2D-SIM data processing workflows.
What are the prospects for further development?
An inherent limitation of all SIM algorithms is the reduced reconstruction quality when processing raw data with degraded illumination pattern quality, due to the shared theoretical basis of structured illumination and demodulation. In the case of Lock-in-SIM, this issue may be further exacerbated due to its increased reliance on the accurate extraction and utilization of a clean grating illumination pattern. By precisely controlling data acquisition quality and further integrating advancements such as adaptive optics, illumination field flatting, and deep learning denoising, the high-fidelity background suppression capability of Lock-in-SIM can be expected to be maintained even under highly challenging experimental conditions.
Where can people find more information?
Methodological details of Lock-in-SIM technique can be found in our publication: Liu W., Zhang M., Zhu W. et al. Visualizing intraorganellar ultrastructures, dynamics, and interactions with open-access background-free Lock-in-SIM. Nature Communications 16, 10765 (2025).
Open-access software can be found in our GitHub: https://github.com/WenjieLab/Lock-in-SIM.
Open-access Lock-in-SIM raw data can be downloaded from our Figshare: https://figshare.com/articles/figure/Open-access_raw_datasets_for_Lock-in-SIM/26130994.


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)