Imaging spotlight: ShapeSpaceExplorer
Posted by FocalPlane, on 5 February 2026
In this paper highlight, we learn about ShapeSpaceExplorer, a tool for measuring cell shape of migrating cells, developed by Samuel Jefferyes, Anne Straube and colleagues.
Can you briefly describe ShapeSpaceExplorer and tell us what’s new about your method?
We designed ShapeSpaceExplorer as a tool to quantitatively measure highly variable cell shapes of migrating cells and analyse shape changes over time. ShapeSpaceExplorer does not rely on any prior knowledge of suitable shape features, but it instead uses a new landmark-free method (which we call best alignment metric) to compute shape differences between pairs of shapes. Diffusion map embedding is used as a non-linear approach for low dimensional representation of shape space, in which similar shapes are close to each other and the local structure in the dataset is preserved. Shape changes are movement through shape space and can therefore be quantified.
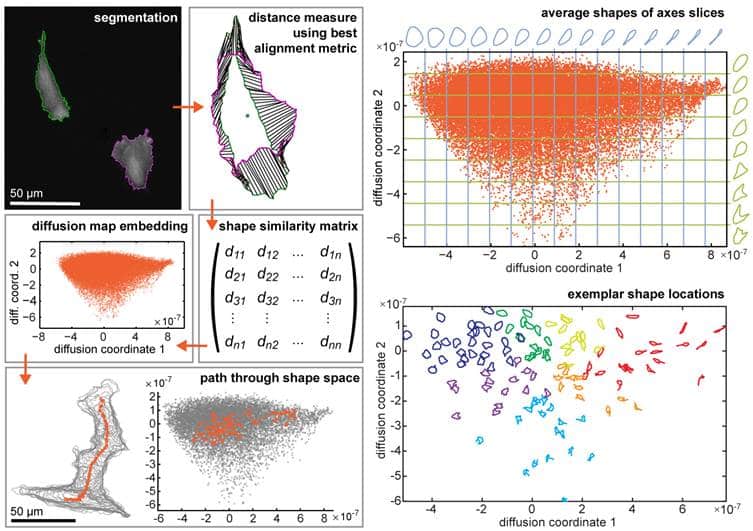
How’s it been used for so far?
We applied ShapeSpaceExplorer to migrating cells and quantified differences in cell morphology distributions across different perturbation experiments. We also measured the speed of cell shape changes globally and between different regions of shape space. For example, we can detect changes in the elongation and retraction speed of cell tails. We also show that shape dynamics information alone can be used to predict when migrating cells change direction. We included various interactive tools for the user to explore shape space, generate average shapes from user-defined areas, partition shape space to analyse distributions of shapes across different experimental conditions. We have also used the software to analyse cell shape changes in response to virus-infection, which works equally well.
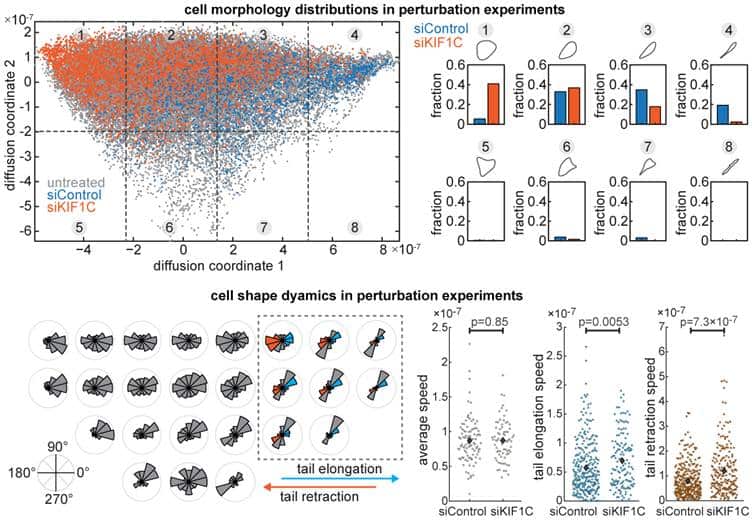
How else could the software be used?
The software works for any set of images from which outlines can be extracted or directly with datasets of 2D closed curves. This could be cells, protein complexes, tissues, organisms, or inanimate objects. While developing the software was motivated by questions in cell biology, the software could find wide applications in science and engineering.
What do researchers require to implement it?
The software is based on MATLAB. We fully tested the latest version with Matlab R2022b and 2024b and we plan to keep it updated. It requires the following MATLAB toolboxes: Image Processing, MATLAB Coder, MATLAB Compiler, Statistics and Machine Learning, (optional) Deep Learning. Detailed documentation with step-by-step instructions and intuitive graphic-user interfaces make the software accessible. No coding experience required.
What are the prospects for further development?
SpaceSpaceExplorer is user-friendly and robust, making cell morphology analysis straightforward and producing various figures and graphs. There is scope for developing advanced analysis tools for shape dynamics. For example to thoroughly correlate shape changes with cell migration or other cell behaviour observed from additional markers in the images.
Where can people find more information?
See the paper:
Samuel D.R. Jefferyes, Roswitha Gostner, Laura Cooper, Mohammed M Abdelsamea, Elly Straube, Nasir Rajpoot, David B.A. Epstein, and Anne Straube (2026). ShapeSpaceExplorer: Analysis of morphological transitions in migrating cells using similarity-based shape space mapping. PLoS Comput Biol 22(1): e1013864. https://doi.org/10.1371/journal.pcbi.1013864
The documentation is here: https://cmcb-warwick.github.io/ShapeSpaceExplorer/
The software is here: https://github.com/cmcb-warwick/ShapeSpaceExplorer Our datasets (raw images and processed data) used are here: https://zenodo.org/records/14998399 and https://zenodo.org/records/14999396


 (No Ratings Yet)
(No Ratings Yet)