Biologists @ 100 conference report
Posted by Margarida Araujo, on 8 May 2025
Biologists @ 100 conference highlights
The Company of Biologists, a not-for-profit organization, plays a pivotal role beyond journal publishing by actively supporting biologists and the broader scientific community. At the recent Biologists @ 100 conference celebrating the 100 years of The Company of Biologists held in Liverpool from 24-27th March 2025, an early-career researcher (ECR) session provided insights into diverse career paths within and outside academia, highlighting roles such as group leader, journal editor, community manager and online editor, research associate in pharmaceutical companies, and project manager in the pharma sector. The event also featured brainstorming and interactive plenary sessions with dedicated Q&A opportunities, fostering dynamic discussions.
A broad spectrum of topics was addressed, ranging from climate change science, offering actionable suggestions for immediate change, to stem cells, developmental biology, cancer, immunology, and cell division. As a first-time attendee as conference reporter, I had the unique chance to attend climate change talks in person for the first time. The conference encouraged ECRs to actively engage by asking questions, while also celebrating exceptional scientists at all career stages through awards for Ph.D. students, postdoctoral researchers, and senior scientists for their contributions to science and mentorship.
The final session explored the intersection of current technologies and future advancements, emphasizing how tools like AI are shaping the present and future of research, from automated image analysis and quantification pipelines to identifying distinct morphology patterns and cell types from existing datasets. Reflecting on this theme, the session resonated with Sydney Brenner’s insight: “Progress in science depends on new techniques, new discoveries, and new ideas, probably in that order.”
Mural created live by the artist Vic Lee during the conference.
Two inspiring climate scientists drew attention to human impacts on climate change and advised taking action now, as the time for mitigation of these impacts is increasingly reduced.
Professor Dr Hans-Otto Pörtner from Alfred Wegener Institut – Climate impacts on animal life and biodiversity: from understanding biogeography limits to the human niche.
Prof. Hans-Otto Pörtner started his talk by identifying the existential threats of climate change and biodiversity loss, as nature and humankind are experiencing changing environments at a planetary scale. Specifically, changing temperatures (global warming) and the distribution of water across the globe are essential factors that impact soil moisture, leading to predicted water shortages in Mediterranean regions in the future. Hans was directly involved in preparing the IPBES-IPCC co-sponsored workshop report, which describes in detail the current imbalance between three interacting systems – climate, biodiversity, and human society – and enumerates pressing and possible solutions that must be implemented. Prof. Hans also showed how Earth’s climate and ecosystems influence each other and define the basis of life for humans. Specifically, Prof. Hans stressed that climate change is causing an impact on the oceans through warming, acidification, and expanding hypoxia in ocean water. Hans emphasized the need to understand the evolutionary background and patterns of biodiversity loss, and reiterated that temperature influences biodiversity in the light of concepts like thermal tolerance and thermal responsiveness. In the IPBES-IPCC report, suggested solutions include mosaic spatial planning and implementation, and conservation of carbon (C-)-rich habitats with enhancement of C-storage. However, we currently face a limited time window to take action and ensure the implementation of these solutions, and the time required for mitigation/maneuvering is increasingly reduced.
Professor Dame Jane Francis from British Antarctic Survey – When Antarctica was green: fossil plants reveal Antarctica’s climate history.
Dame Jane Francis started by presenting beautiful pictures from her field expeditions of present-day icy Antarctica, but reminded the audience that Antarctica was not always a land of ice! 100 million years ago (Cretaceous age), the Antarctica continent was situated over the South Pole and was covered in forests and dinosaurs. Dame Jane Francis provided evidence of the past environmental conditions and Antarctic forests coming from pollen, leaves, and fossil wood. Among these fossils are astoundingly well-conserved, petrified tree trunks exhibiting tree rings in trunk sections, and monkey puzzle trees, belonging mostly to tree ferns and conifers. Interestingly, monkey puzzle trees can be found today in the Andes, Chile, and many Antarctic plants are ancestors of most present-day southern hemisphere vegetation in South America, New Zealand, and Australia. After careful in situ mapping of petrified tree stumps, roots, and leaves by a Ph.D. student and in collaboration with an artist, Bob Nichols, they generated a reconstructed image depicting the environment and flora of Antarctica during the Cretaceous period, driving the audience’s imagination and transporting it into those forests. There is even a hidden dinosaur in the reconstructed image, which Dame Jane challenged the audience to find!
“The onset of the ice house”.
As tectonic plates moved apart, Antarctica became separated from other continents and was isolated in cold, icy waters. How is climate change affecting Antarctica? How is climate warming affecting ice sheets? Antarctica as we know it today is covered by big ice streams and, currently, ice shelves are melting from below due to progressively warmer seas, leading to the rise of the sea level. Dame Jane ended her talk by sharing a visualization tool – NASA Scientific Visualization Tool Studio, which produces visualizations, animations, and images to promote a greater understanding of Earth and Space Sciences.
Exciting talks showcasing new imaging techniques
As a conference reporter for FocalPlane, the community site for researchers who use microscopy in their work, I chose to highlight two galvanizing talks where both cell biology and microscopy are the main actors. One talk is about centrosome biogenesis from Dr Jordan Raff (University of Oxford), and the other is about protein translation on the ER and emerging tools in cell biology research from Dr Jennifer Lippincott-Schwartz (Janelia Research Campus of the Howard Hughes Medical Institute). These talks showcase how revolutionary imaging techniques are enhancing our understanding of cell internal structures and dynamic processes, advancing cell biology and biomedical research.
Dr Jordan Raff – University of Oxford
Reconstituting organelle biogenesis on synthetic structures.
Dr Jordan Raff started his talk by referring to the diverse array of organelles present in eukaryotic cells, reinforcing that their numbers and dimensions must be stringently regulated. In particular, in dividing cells, organelle content generally doubles before cell division; however, the mechanisms governing the assembly, growth, and coordination of these complex structures remain unresolved questions in the field. To address these questions, Jordan ’s lab focuses on centrosome/centriole assembly as a model to understand the principles governing organelle biogenesis in space and time. Jordan reminded the audience that centrosomes duplicate precisely once per cell cycle and that these membrane-less organelles comprise a central pair of centrioles surrounded by a matrix of pericentriolar material (PCM). In their recent inspiring experiments, Jordan ’s team reconstituted centrosome biogenesis by employing micron-sized synthetic beads injected into early Drosophila embryos, as seen in the following figure.
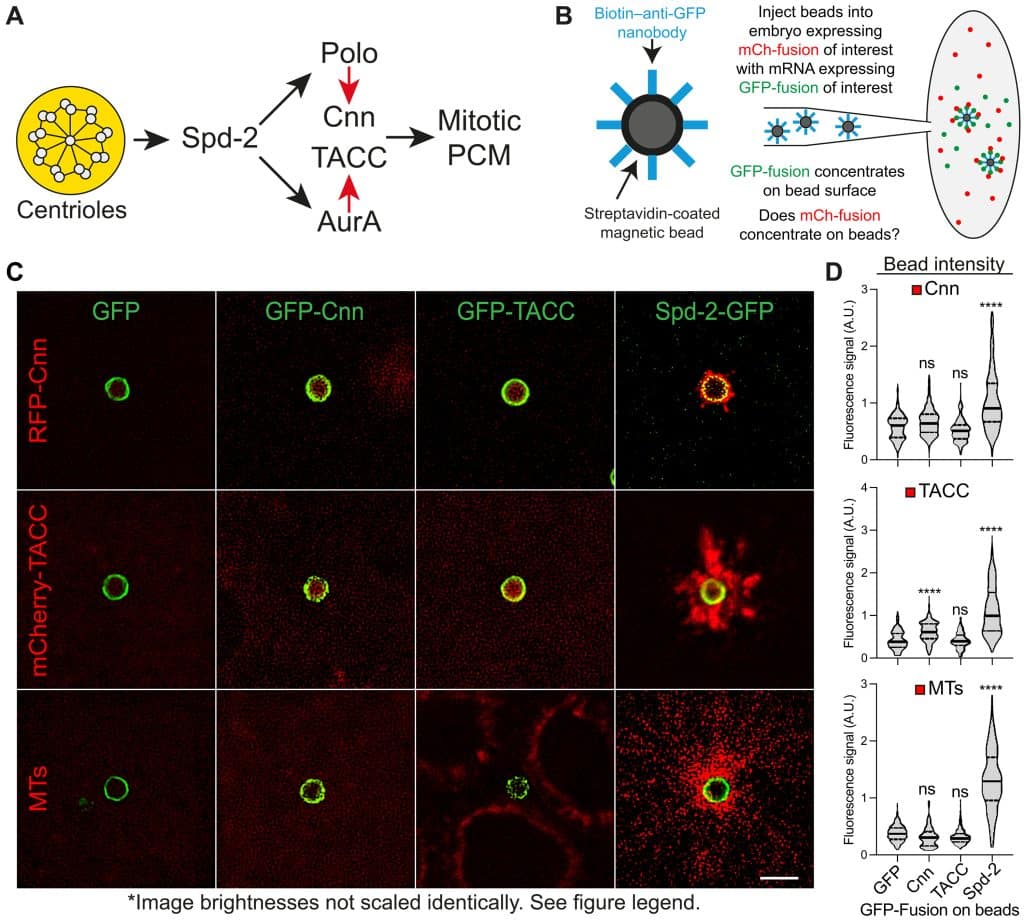
Remarkably, these micron-size beads that are functionalized (surface-coated) to recruit individual core centriole assembly proteins lead to the formation of centrioles closely mimicking endogenous structures. Jordan showed live imaging movies where synthetic centrioles spectacularly bud off/dissociate from the bead surface. These experiments revealed that just like endogenous centrioles, these synthetic centrioles also become regularly spaced, can assemble PCM, and undergo successive rounds of duplication in synchrony with the embryonic cell cycle. Jordan went on to show that centrioles generate an outward flux of Spd-2 molecules and that recruiting Spd-2, but not Cnn or TACC, to the surface of synthetic beads injected into early embryos is sufficient to reconstitute several aspects of mitotic PCM assembly on the bead surface. These recent findings from Jordan ’s team point to a conserved and surprisingly simple ‘core’ pathway of centriole assembly. This provides significant insights into the fundamental principles underlying centriole biogenesis and suggests a remarkable capacity for synthetic systems to mimic cellular processes with high precision.
Reference: Siu-Shing Wong et al., Centrioles generate two scaffolds with distinct biophysical properties to build mitotic centrosomes. Sci. Adv. 11, eadq9549 (2025). doi:10.1126/sciadv.adq9549.
Dr Jennifer Lippincott-Schwartz – Group Leader at the Janelia Research Campus of the Howard Hughes Medical Institute
Powerful new ways to image the internal structures and complex dynamics of cells are revolutionizing cell biology and biomedical research.
Dr Jennifer Lippincott-Schwartz is a cell biologist whose pioneering work has significantly advanced our understanding of intracellular dynamics and organelle organization. In her inspiring talk, Jennifer z explored how to image protein synthesis at a micron scale, focusing on the core machine of translation, the ribosome. While micron-sized ribosomes are found throughout the cell in Archaea, eukaryotes have ER-bound ribosomes that translate membrane and secretory proteins, involving co-translational insertion into ER membranes. Jennifer showed beautiful confocal microscopy images depicting the incredibly complex and diverse morphologies of the largest organelle within the cell: the ER. But where, when, and how does protein synthesis occur within the ER? To address this question, Jennifer’s team tagged mRNAs with MS2 reporter binding sites, in particular, ‘secretome’” mRNAs encoding membrane or secretory proteins, which are co-translationally translocated into the ER membrane. Using this method, Jennifer’s team identified two distinct types of mRNAs according to their motion, a slow-moving and a fast-moving type. To test which of these different types of movements correlated with MS2 mRNA being actively engaged in co-translational translocation, Jennifer’s team treated cells with puromycin, which terminates translation. This strikingly changed most MS2 reporter mRNAs into a fast-moving population with many mRNAs being displaced from the ER, suggesting that slow-moving mRNA molecules were undergoing translation. Then, Jennifer’s team generated an additional mRNA sensor using a SUNTAG translation reporter. Interestingly, using the SUNTAG translation reporter, they found that the majority of the translation sites are located at particular sites on the ER, specifically at 3-way junctions. To get further structural insight into these particular translation hot spots at 3-way junctions, Jennifer’s team decided to perform a volumetric analysis using FIB-SEM. This detailed volumetric analysis further revealed an intriguing ribosome organization, with ribosomes concentrated at ER junctions and organized in a spiral arrangement, forming chains called polysomes. This raised the question of what exactly makes ER junctions hot spots for translation and whether other organelles could facilitate the process or could also be involved. Jennifer then alluded that lysosomes frequently localize near ER junctions, suggesting that lysosomes could help regulate mRNA translation at these sites. This was confirmed by the observation that lysosomes co-localize with the SUNTAG translation reporter and by treatment with chloroquine, which inhibits lysosome function, reducing translation. Lunapark is a protein enriched at ER junctions, and proximity ligation assays to detect protein-protein interactions revealed enhanced ligation of Lunapark and the lysosome marker LAMP1. Knockdown of Lunapark diminishes SUNTAG intensity near lysosomes, leading to a loss of enhanced translation efficiency. Importantly, knock-out of Lunapark leads to elevated levels of phosphorylated eIF2A, linked to translation initiation control. In addition, FRAP assays further revealed that the recovery of SUNTAG fluorescence intensity is not complete upon Lunapark knockdown. These findings led Jennifer to propose a model in which lysosomes and Lunapark locally pattern and regulate the synthesis of membrane and secretory proteins of the ER. In the final minutes of her talk, Jennifer briefly shared her most recent work leveraging cutting-edge imaging technologies to visualize protein synthesis at unprecedented atomic resolution. This includes real-time tracking of ribosome function, translational machinery dynamics, and spatial regulation of protein production within cells.
Reference: Choi et al., Lysosomal release of amino acids at ER three-way junctions regulates transmembrane and secretory protein mRNA translation. bioRxiv 2023.08.01.551382; doi: https://doi.org/10.1101/2023.08.01.551382.
For researcher interviews, check out part 2 of my conference report: https://focalplane.biologists.com/2025/05/08/interviews-and-researchers-testimonies-from-biologists-100/


 (No Ratings Yet)
(No Ratings Yet)